Modeling the effect of codon translation rates on co-translational protein folding mechanisms of arbitrary complexity
- PMID: 25877595
- PMCID: PMC6597233
- DOI: 10.1063/1.4916914
Modeling the effect of codon translation rates on co-translational protein folding mechanisms of arbitrary complexity
Abstract
In a cell, the folding of a protein molecule into tertiary structure can begin while it is synthesized by the ribosome. The rate at which individual amino acids are incorporated into the elongating nascent chain has been shown to affect the likelihood that proteins will populate their folded state, indicating that co-translational protein folding is a far from equilibrium process. Developing a theoretical framework to accurately describe this process is, therefore, crucial for advancing our understanding of how proteins acquire their functional conformation in living cells. Current state-of-the-art computational approaches, such as molecular dynamics simulations, are very demanding in terms of the required computer resources, making the simulation of co-translational protein folding difficult. Here, we overcome this limitation by introducing an efficient approach that predicts the effects that variable codon translation rates have on co-translational folding pathways. Our approach is based on Markov chains. By using as an input a relatively small number of molecular dynamics simulations, it allows for the computation of the probability that a nascent protein is in any state as a function of the translation rate of individual codons along a mRNA's open reading frame. Due to its computational efficiency and favorable scalability with the complexity of the folding mechanism, this approach could enable proteome-wide computational studies of the influence of translation dynamics on co-translational folding.
Figures
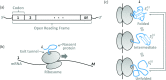
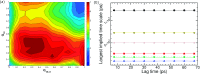
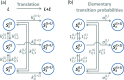
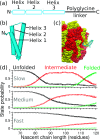
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

