The Ubp15 deubiquitinase promotes timely entry into S phase in Saccharomyces cerevisiae
- PMID: 25877870
- PMCID: PMC4462939
- DOI: 10.1091/mbc.E14-09-1400
The Ubp15 deubiquitinase promotes timely entry into S phase in Saccharomyces cerevisiae
Abstract
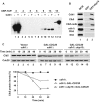
The anaphase-promoting complex in partnership with its activator, Cdh1, is an E3 ubiquitin ligase responsible for targeting cell cycle proteins during G1 phase. In the budding yeast Saccharomyces cerevisiae, Cdh1 associates with the deubiquitinating enzyme Ubp15, but the significance of this interaction is unclear. To better understand the physiological role(s) of Ubp15, we examined cell cycle phenotypes of cells lacking Ubp15. We found that ubp15∆ cells exhibited delayed progression from G1 into S phase and increased sensitivity to the DNA synthesis inhibitor hydroxyurea. Both phenotypes of ubp15∆ cells were rescued by additional copies of the S-phase cyclin gene CLB5. Clb5 is an unstable protein targeted for proteasome-mediated degradation by several pathways. We found that during G1 phase, the APC(Cdh1)-mediated degradation of Clb5 was accelerated in ubp15∆ cells. Ubp15 interacted with Clb5 independent of Cdh1 and deubiquitinated Clb5 in a reconstituted system. Thus deubiquitination by Ubp15 counteracts APC activity toward cyclin Clb5 to allow Clb5 accumulation and a timely entry into S phase.
© 2015 Ostapenko et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures






Similar articles
-
A process independent of the anaphase-promoting complex contributes to instability of the yeast S phase cyclin Clb5.J Biol Chem. 2007 Sep 7;282(36):26614-22. doi: 10.1074/jbc.M703744200. Epub 2007 Jul 9. J Biol Chem. 2007. PMID: 17620341
-
APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5.Nature. 1999 Nov 11;402(6758):203-7. doi: 10.1038/46080. Nature. 1999. PMID: 10647015
-
Phosphorylation and dephosphorylation regulate APC/C(Cdh1) substrate degradation.Cell Cycle. 2015;14(19):3138-45. doi: 10.1080/15384101.2015.1078036. Cell Cycle. 2015. PMID: 26252546 Free PMC article.
-
S-phase cyclin-dependent kinases promote sister chromatid cohesion in budding yeast.Mol Cell Biol. 2011 Jun;31(12):2470-83. doi: 10.1128/MCB.05323-11. Epub 2011 Apr 25. Mol Cell Biol. 2011. PMID: 21518961 Free PMC article.
-
An APC/C-Cdh1 Biosensor Reveals the Dynamics of Cdh1 Inactivation at the G1/S Transition.PLoS One. 2016 Jul 13;11(7):e0159166. doi: 10.1371/journal.pone.0159166. eCollection 2016. PLoS One. 2016. PMID: 27410035 Free PMC article.
Cited by
-
Cezanne/OTUD7B is a cell cycle-regulated deubiquitinase that antagonizes the degradation of APC/C substrates.EMBO J. 2018 Aug 15;37(16):e98701. doi: 10.15252/embj.201798701. Epub 2018 Jul 4. EMBO J. 2018. PMID: 29973362 Free PMC article.
-
Protein Homeostasis Networks and the Use of Yeast to Guide Interventions in Alzheimer's Disease.Int J Mol Sci. 2020 Oct 28;21(21):8014. doi: 10.3390/ijms21218014. Int J Mol Sci. 2020. PMID: 33126501 Free PMC article. Review.
-
Insights into APC/C: from cellular function to diseases and therapeutics.Cell Div. 2016 Jul 13;11:9. doi: 10.1186/s13008-016-0021-6. eCollection 2016. Cell Div. 2016. PMID: 27418942 Free PMC article. Review.
-
Genetic analysis reveals functions of atypical polyubiquitin chains.Elife. 2018 Dec 14;7:e42955. doi: 10.7554/eLife.42955. Elife. 2018. PMID: 30547882 Free PMC article.
-
Recent Progress of Deubiquitinating Enzymes in Human and Plant Pathogenic Fungi.Biomolecules. 2022 Oct 4;12(10):1424. doi: 10.3390/biom12101424. Biomolecules. 2022. PMID: 36291632 Free PMC article. Review.
References
-
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. - PubMed
-
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. - PubMed
-
- Bozza WP, Zhuang Z. Biochemical characterization of a multidomain deubiquitinating enzyme Ubp15 and the regulatory role of its terminal domains. Biochemistry. 2011;50:6423–6432. - PubMed
-
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

