Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
- PMID: 25877892
- PMCID: PMC4636056
- DOI: 10.1126/scitranslmed.aaa1408
Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
Abstract
Deciphering whether actionable driver mutations are found in all or a subset of tumor cells will likely be required to improve drug development and precision medicine strategies. We analyzed nine cancer types to determine the subclonal frequencies of driver events, to time mutational processes during cancer evolution, and to identify drivers of subclonal expansions. Although mutations in known driver genes typically occurred early in cancer evolution, we also identified later subclonal "actionable" mutations, including BRAF (V600E), IDH1 (R132H), PIK3CA (E545K), EGFR (L858R), and KRAS (G12D), which may compromise the efficacy of targeted therapy approaches. More than 20% of IDH1 mutations in glioblastomas, and 15% of mutations in genes in the PI3K (phosphatidylinositol 3-kinase)-AKT-mTOR (mammalian target of rapamycin) signaling axis across all tumor types were subclonal. Mutations in the RAS-MEK (mitogen-activated protein kinase kinase) signaling axis were less likely to be subclonal than mutations in genes associated with PI3K-AKT-mTOR signaling. Analysis of late mutations revealed a link between APOBEC-mediated mutagenesis and the acquisition of subclonal driver mutations and uncovered putative cancer genes involved in subclonal expansions, including CTNNA2 and ATXN1. Our results provide a pan-cancer census of driver events within the context of intratumor heterogeneity and reveal patterns of tumor evolution across cancers. The frequent presence of subclonal driver mutations suggests the need to stratify targeted therapy response according to the proportion of tumor cells in which the driver is identified.
Copyright © 2015, American Association for the Advancement of Science.
Figures

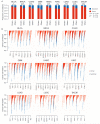
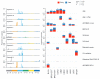
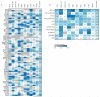
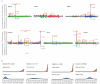
Comment in
-
Genetics: clonal and subclonal events in cancer evolution--optimizing cancer therapy.Nat Rev Clin Oncol. 2015 Jul;12(7):372. doi: 10.1038/nrclinonc.2015.87. Epub 2015 May 5. Nat Rev Clin Oncol. 2015. PMID: 25940986 No abstract available.
References
-
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. - PMC - PubMed
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, VanBuuren MM, van ’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

