Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae
- PMID: 25878099
- PMCID: PMC4468488
- DOI: 10.1128/JVI.00115-15
Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae
Abstract
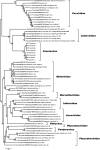
Giant viruses are protist-associated viruses belonging to the proposed order Megavirales; almost all have been isolated from Acanthamoeba spp. Their isolation in humans suggests that they are part of the human virome. Using a high-throughput strategy to isolate new giant viruses from their original protozoan hosts, we obtained eight isolates of a new giant viral lineage from Vermamoeba vermiformis, the most common free-living protist found in human environments. This new lineage was proposed to be the faustovirus lineage. The prototype member, faustovirus E12, forms icosahedral virions of ≈ 200 nm that are devoid of fibrils and that encapsidate a 466-kbp genome encoding 451 predicted proteins. Of these, 164 are found in the virion. Phylogenetic analysis of the core viral genes showed that faustovirus is distantly related to the mammalian pathogen African swine fever virus, but it encodes ≈ 3 times more mosaic gene complements. About two-thirds of these genes do not show significant similarity to genes encoding any known proteins. These findings show that expanding the panel of protists to discover new giant viruses is a fruitful strategy.
Importance: By using Vermamoeba, a protist living in humans and their environment, we isolated eight strains of a new giant virus that we named faustovirus. The genomes of these strains were sequenced, and their sequences showed that faustoviruses are related to but different from the vertebrate pathogen African swine fever virus (ASFV), which belongs to the family Asfarviridae. Moreover, the faustovirus gene repertoire is ≈ 3 times larger than that of ASFV and comprises approximately two-thirds ORFans (open reading frames [ORFs] with no detectable homology to other ORFs in a database).
Figures


References
-
- Colson P, de Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521. doi:10.1007/s00705-013-1768-6. - DOI - PMC - PubMed
-
- Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi:10.1126/science.1239181. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

