Glycan Microheterogeneity at the PGT135 Antibody Recognition Site on HIV-1 gp120 Reveals a Molecular Mechanism for Neutralization Resistance
- PMID: 25878100
- PMCID: PMC4468474
- DOI: 10.1128/JVI.00230-15
Glycan Microheterogeneity at the PGT135 Antibody Recognition Site on HIV-1 gp120 Reveals a Molecular Mechanism for Neutralization Resistance
Abstract
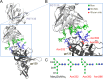
Broadly neutralizing antibodies have been isolated that bind the glycan shield of the HIV-1 envelope spike. One such antibody, PGT135, contacts the intrinsic mannose patch of gp120 at the Asn332, Asn392, and Asn386 glycosylation sites. Here, site-specific glycosylation analysis of recombinant gp120 revealed glycan microheterogeneity sufficient to explain the existence of a minor population of virions resistant to PGT135 neutralization. Target microheterogeneity and antibody glycan specificity are therefore important parameters in HIV-1 vaccine design.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Conserved Role of an N-Linked Glycan on the Surface Antigen of Human Immunodeficiency Virus Type 1 Modulating Virus Sensitivity to Broadly Neutralizing Antibodies against the Receptor and Coreceptor Binding Sites.J Virol. 2015 Oct 28;90(2):829-41. doi: 10.1128/JVI.02321-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26512079 Free PMC article.
-
Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition.Virology. 2003 Sep 1;313(2):387-400. doi: 10.1016/s0042-6822(03)00294-0. Virology. 2003. PMID: 12954207
-
Chinks in the armor of the HIV-1 Envelope glycan shield: Implications for immune escape from anti-glycan broadly neutralizing antibodies.Virology. 2017 Jan 15;501:12-24. doi: 10.1016/j.virol.2016.10.026. Epub 2016 Nov 13. Virology. 2017. PMID: 27846415
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
-
GP120: target for neutralizing HIV-1 antibodies.Annu Rev Immunol. 2006;24:739-69. doi: 10.1146/annurev.immunol.24.021605.090557. Annu Rev Immunol. 2006. PMID: 16551265 Review.
Cited by
-
Global site-specific N-glycosylation analysis of HIV envelope glycoprotein.Nat Commun. 2017 Mar 28;8:14954. doi: 10.1038/ncomms14954. Nat Commun. 2017. PMID: 28348411 Free PMC article.
-
The HIV glycan shield as a target for broadly neutralizing antibodies.FEBS J. 2015 Dec;282(24):4679-91. doi: 10.1111/febs.13530. Epub 2015 Oct 23. FEBS J. 2015. PMID: 26411545 Free PMC article. Review.
-
The HIV-1 envelope glycoprotein: structure, function and interactions with neutralizing antibodies.Nat Rev Microbiol. 2025 Jul 23. doi: 10.1038/s41579-025-01206-6. Online ahead of print. Nat Rev Microbiol. 2025. PMID: 40702326 Review.
-
Structural Rearrangements Maintain the Glycan Shield of an HIV-1 Envelope Trimer After the Loss of a Glycan.Sci Rep. 2018 Oct 9;8(1):15031. doi: 10.1038/s41598-018-33390-2. Sci Rep. 2018. PMID: 30302011 Free PMC article.
-
Antibody recognition of HIV and dengue glycoproteins.Glycobiology. 2016 Aug;26(8):813-9. doi: 10.1093/glycob/cww031. Epub 2016 Mar 3. Glycobiology. 2016. PMID: 26941393 Free PMC article. Review.
References
-
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi:10.1126/science.1178746. - DOI - PMC - PubMed
-
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi:10.1038/nature10373. - DOI - PMC - PubMed
-
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang C-H, Mcbride R, von Bredow B, Shivatare SS, Wu C-Y, Chan-Hui P-Y, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong C-H, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi:10.1016/j.immuni.2014.04.009. - DOI - PMC - PubMed
-
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. doi:10.1016/j.chom.2012.09.008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

