The Vesicle-Forming 6K2 Protein of Turnip Mosaic Virus Interacts with the COPII Coatomer Sec24a for Viral Systemic Infection
- PMID: 25878114
- PMCID: PMC4468490
- DOI: 10.1128/JVI.00503-15
The Vesicle-Forming 6K2 Protein of Turnip Mosaic Virus Interacts with the COPII Coatomer Sec24a for Viral Systemic Infection
Abstract
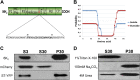
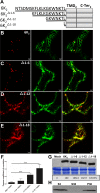
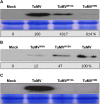
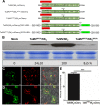
Positive-sense RNA viruses remodel host cell endomembranes to generate quasi-organelles known as "viral factories" to coordinate diverse viral processes, such as genome translation and replication. It is also becoming clear that enclosing viral RNA (vRNA) complexes within membranous structures is important for virus cell-to-cell spread throughout the host. In plant cells infected by turnip mosaic virus (TuMV), a member of the family Potyviridae, peripheral motile endoplasmic reticulum (ER)-derived viral vesicles are produced that carry the vRNA to plasmodesmata for delivery into adjacent noninfected cells. The viral protein 6K2 is responsible for the formation of these vesicles, but how 6K2 is involved in their biogenesis is unknown. We show here that 6K2 is associated with cellular membranes. Deletion mapping and site-directed mutagenesis experiments defined a soluble N-terminal 12-amino-acid stretch, in particular a potyviral highly conserved tryptophan residue and two lysine residues that were important for vesicle formation. When the tryptophan residue was changed into an alanine in the viral polyprotein, virus replication still took place, albeit at a reduced level, but cell-to-cell movement of the virus was abolished. Yeast (Saccharomyces cerevisiae) two-hybrid and coimmunoprecipitation experiments showed that 6K2 interacted with Sec24a, a COPII coatomer component. Appropriately, TuMV systemic movement was delayed in an Arabidopsis thaliana mutant line defective in Sec24a. Intercellular movement of TuMV replication vesicles thus requires ER export of 6K2, which is mediated by the interaction of the N-terminal domain of the viral protein with Sec24a.
Importance: Many plant viruses remodel the endoplasmic reticulum (ER) to generate vesicles that are associated with the virus replication complex. The viral protein 6K2 of turnip mosaic virus (TuMV) is known to induce ER-derived vesicles that contain vRNA as well as viral and host proteins required for vRNA synthesis. These vesicles not only sustain vRNA synthesis, they are also involved in the intercellular trafficking of vRNA. In this investigation, we found that the N-terminal soluble domain of 6K2 is required for ER export of the protein and for the formation of vesicles. ER export is not absolutely required for vRNA replication but is necessary for virus cell-to-cell movement. Furthermore, we found that 6K2 physically interacts with the COPII coatomer Sec24a and that an Arabidopsis thaliana mutant line with a defective Sec24a shows a delay in the systemic infection by TuMV.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Turnip Mosaic Virus Uses the SNARE Protein VTI11 in an Unconventional Route for Replication Vesicle Trafficking.Plant Cell. 2018 Oct;30(10):2594-2615. doi: 10.1105/tpc.18.00281. Epub 2018 Aug 27. Plant Cell. 2018. PMID: 30150314 Free PMC article.
-
Two plant membrane-shaping reticulon-like proteins play contrasting complex roles in turnip mosaic virus infection.Mol Plant Pathol. 2024 Oct;25(10):e70017. doi: 10.1111/mpp.70017. Mol Plant Pathol. 2024. PMID: 39412487 Free PMC article.
-
A Host ER Fusogen Is Recruited by Turnip Mosaic Virus for Maturation of Viral Replication Vesicles.Plant Physiol. 2019 Feb;179(2):507-518. doi: 10.1104/pp.18.01342. Epub 2018 Dec 11. Plant Physiol. 2019. PMID: 30538165 Free PMC article.
-
Plant virus replication and movement.Virology. 2015 May;479-480:657-71. doi: 10.1016/j.virol.2015.01.025. Epub 2015 Mar 3. Virology. 2015. PMID: 25746797 Review.
-
The Molecular Maze of Potyviral and Host Protein Interactions.Annu Rev Virol. 2024 Sep;11(1):147-170. doi: 10.1146/annurev-virology-100422-034124. Epub 2024 Aug 30. Annu Rev Virol. 2024. PMID: 38848589 Review.
Cited by
-
A novel block of plant virus movement genes.Mol Plant Pathol. 2017 Jun;18(5):611-624. doi: 10.1111/mpp.12418. Epub 2016 Jul 15. Mol Plant Pathol. 2017. PMID: 27118327 Free PMC article.
-
An engineered mutant of a host phospholipid synthesis gene inhibits viral replication without compromising host fitness.J Biol Chem. 2019 Sep 20;294(38):13973-13982. doi: 10.1074/jbc.RA118.007051. Epub 2019 Jul 30. J Biol Chem. 2019. PMID: 31362985 Free PMC article.
-
Salicylic acid and RNA interference mediate antiviral immunity of plant stem cells.Proc Natl Acad Sci U S A. 2023 Oct 17;120(42):e2302069120. doi: 10.1073/pnas.2302069120. Epub 2023 Oct 12. Proc Natl Acad Sci U S A. 2023. PMID: 37824524 Free PMC article.
-
Distinct Mechanisms of Endomembrane Reorganization Determine Dissimilar Transport Pathways in Plant RNA Viruses.Plants (Basel). 2022 Sep 15;11(18):2403. doi: 10.3390/plants11182403. Plants (Basel). 2022. PMID: 36145804 Free PMC article. Review.
-
Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes.AIMS Microbiol. 2020 Sep 21;6(3):305-329. doi: 10.3934/microbiol.2020019. eCollection 2020. AIMS Microbiol. 2020. PMID: 33134746 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

