Immunogenic Display of Purified Chemically Cross-Linked HIV-1 Spikes
- PMID: 25878116
- PMCID: PMC4468504
- DOI: 10.1128/JVI.03738-14
Immunogenic Display of Purified Chemically Cross-Linked HIV-1 Spikes
Abstract
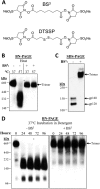
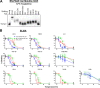
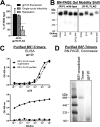
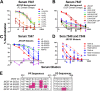
HIV-1 envelope glycoprotein (Env) spikes are prime vaccine candidates, at least in principle, but suffer from instability, molecular heterogeneity and a low copy number on virions. We anticipated that chemical cross-linking of HIV-1 would allow purification and molecular characterization of trimeric Env spikes, as well as high copy number immunization. Broadly neutralizing antibodies bound tightly to all major quaternary epitopes on cross-linked spikes. Covalent cross-linking of the trimer also stabilized broadly neutralizing epitopes, although surprisingly some individual epitopes were still somewhat sensitive to heat or reducing agent. Immunodepletion using non-neutralizing antibodies to gp120 and gp41 was an effective method for removing non-native-like Env. Cross-linked spikes, purified via an engineered C-terminal tag, were shown by negative stain EM to have well-ordered, trilobed structure. An immunization was performed comparing a boost with Env spikes on virions to spikes cross-linked and captured onto nanoparticles, each following a gp160 DNA prime. Although differences in neutralization did not reach statistical significance, cross-linked Env spikes elicited a more diverse and sporadically neutralizing antibody response against Tier 1b and 2 isolates when displayed on nanoparticles, despite attenuated binding titers to gp120 and V3 crown peptides. Our study demonstrates display of cross-linked trimeric Env spikes on nanoparticles, while showing a level of control over antigenicity, purity and density of virion-associated Env, which may have relevance for Env based vaccine strategies for HIV-1.
Importance: The envelope spike (Env) is the target of HIV-1 neutralizing antibodies, which a successful vaccine will need to elicit. However, native Env on virions is innately labile, as well as heterogeneously and sparsely displayed. We therefore stabilized Env spikes using a chemical cross-linker and removed non-native Env by immunodepletion with non-neutralizing antibodies. Fixed native spikes were recognized by all classes of known broadly neutralizing antibodies but not by non-neutralizing antibodies and displayed on nanoparticles in high copy number. An immunization experiment in rabbits revealed that cross-linking Env reduced its overall immunogenicity; however, high-copy display on nanoparticles enabled boosting of antibodies that sporadically neutralized some relatively resistant HIV-1 isolates, albeit at a low titer. This study describes the purification of stable and antigenically correct Env spikes from virions that can be used as immunogens.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures









Similar articles
-
Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins.PLoS Pathog. 2018 May 10;14(5):e1006986. doi: 10.1371/journal.ppat.1006986. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29746590 Free PMC article.
-
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.J Virol. 2018 Feb 12;92(5):e01796-17. doi: 10.1128/JVI.01796-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237847 Free PMC article.
-
Immunogenicity of a Prefusion HIV-1 Envelope Trimer in Complex with a Quaternary-Structure-Specific Antibody.J Virol. 2015 Dec 30;90(6):2740-55. doi: 10.1128/JVI.02380-15. J Virol. 2015. PMID: 26719262 Free PMC article.
-
Challenges for structure-based HIV vaccine design.Curr Opin HIV AIDS. 2009 Sep;4(5):431-40. doi: 10.1097/COH.0b013e32832e6184. Curr Opin HIV AIDS. 2009. PMID: 20048708 Review.
-
Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies.Retrovirology. 2018 Sep 12;15(1):63. doi: 10.1186/s12977-018-0445-y. Retrovirology. 2018. PMID: 30208933 Free PMC article. Review.
Cited by
-
Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins.PLoS Pathog. 2018 May 10;14(5):e1006986. doi: 10.1371/journal.ppat.1006986. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29746590 Free PMC article.
-
Conformations of Human Immunodeficiency Virus Envelope Glycoproteins in Detergents and Styrene-Maleic Acid Lipid Particles.J Virol. 2023 Jun 29;97(6):e0032723. doi: 10.1128/jvi.00327-23. Epub 2023 May 31. J Virol. 2023. PMID: 37255444 Free PMC article.
-
Effects of partially dismantling the CD4 binding site glycan fence of HIV-1 Envelope glycoprotein trimers on neutralizing antibody induction.Virology. 2017 May;505:193-209. doi: 10.1016/j.virol.2017.02.024. Epub 2017 Mar 6. Virology. 2017. PMID: 28279830 Free PMC article.
-
Artificial Anti-HIV-1 Immunogen Comprising Epitopes of Broadly Neutralizing Antibodies 2F5, 10E8, and a Peptide Mimic of VRC01 Discontinuous Epitope.Vaccines (Basel). 2019 Aug 6;7(3):83. doi: 10.3390/vaccines7030083. Vaccines (Basel). 2019. PMID: 31390770 Free PMC article.
-
Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10.J Virol. 2015 Dec;89(23):11975-89. doi: 10.1128/JVI.01793-15. Epub 2015 Sep 16. J Virol. 2015. PMID: 26378169 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

