The cell biology of lignification in higher plants
- PMID: 25878140
- PMCID: PMC4648457
- DOI: 10.1093/aob/mcv046
The cell biology of lignification in higher plants
Abstract
Background: Lignin is a polyphenolic polymer that strengthens and waterproofs the cell wall of specialized plant cell types. Lignification is part of the normal differentiation programme and functioning of specific cell types, but can also be triggered as a response to various biotic and abiotic stresses in cells that would not otherwise be lignifying.
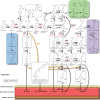
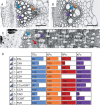
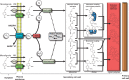
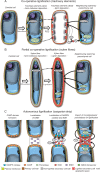
Scope: Cell wall lignification exhibits specific characteristics depending on the cell type being considered. These characteristics include the timing of lignification during cell differentiation, the palette of associated enzymes and substrates, the sub-cellular deposition sites, the monomeric composition and the cellular autonomy for lignin monomer production. This review provides an overview of the current understanding of lignin biosynthesis and polymerization at the cell biology level.
Conclusions: The lignification process ranges from full autonomy to complete co-operation depending on the cell type. The different roles of lignin for the function of each specific plant cell type are clearly illustrated by the multiple phenotypic defects exhibited by knock-out mutants in lignin synthesis, which may explain why no general mechanism for lignification has yet been defined. The range of phenotypic effects observed include altered xylem sap transport, loss of mechanical support, reduced seed protection and dispersion, and/or increased pest and disease susceptibility.
Keywords: Arabidopsis thaliana; Lignin; laccases; lignification; monolignols; non-cell autonomous processes; peroxidases; plant cell wall.
© The Author 2015. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Adler E. 1977. Lignin chemistry – past, present and future. Wood Science and Technology 3: 169–218.
-
- Alejandro S, Lee Y, Tohge T, et al. 2012. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Current Biology 22: 1207–1212. - PubMed
-
- Bao W, O’Malley DM, Whetten R, Sederoff RR. 1993. A laccase associated with lignification in loblolly pine xylem. Science 260: 672–674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

