Heterogeneous Nuclear Ribonucleoprotein C Proteins Interact with the Human Papillomavirus Type 16 (HPV16) Early 3'-Untranslated Region and Alleviate Suppression of HPV16 Late L1 mRNA Splicing
- PMID: 25878250
- PMCID: PMC4505585
- DOI: 10.1074/jbc.M115.638098
Heterogeneous Nuclear Ribonucleoprotein C Proteins Interact with the Human Papillomavirus Type 16 (HPV16) Early 3'-Untranslated Region and Alleviate Suppression of HPV16 Late L1 mRNA Splicing
Abstract
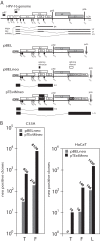
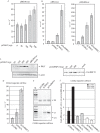
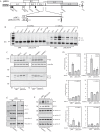
In order to identify cellular factors that regulate human papillomavirus type 16 (HPV16) gene expression, cervical cancer cells permissive for HPV16 late gene expression were identified and characterized. These cells either contained a novel spliced variant of the L1 mRNAs that bypassed the suppressed HPV16 late, 5'-splice site SD3632; produced elevated levels of RNA-binding proteins SRSF1 (ASF/SF2), SRSF9 (SRp30c), and HuR that are known to regulate HPV16 late gene expression; or were shown by a gene expression array analysis to overexpress the RALYL RNA-binding protein of the heterogeneous nuclear ribonucleoprotein C (hnRNP C) family. Overexpression of RALYL or hnRNP C1 induced HPV16 late gene expression from HPV16 subgenomic plasmids and from episomal forms of the full-length HPV16 genome. This induction was dependent on the HPV16 early untranslated region. Binding of hnRNP C1 to the HPV16 early, untranslated region activated HPV16 late 5'-splice site SD3632 and resulted in production of HPV16 L1 mRNAs. Our results suggested that hnRNP C1 controls HPV16 late gene expression.
Keywords: DNA viruses; HPV; RNA processing; RNA splicing; RNA-binding protein; tumor virus.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures








Similar articles
-
Suppression of HPV-16 late L1 5'-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs.Nucleic Acids Res. 2013 Dec;41(22):10488-508. doi: 10.1093/nar/gkt803. Epub 2013 Sep 5. Nucleic Acids Res. 2013. PMID: 24013563 Free PMC article.
-
Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene.J Virol. 2004 Oct;78(20):10888-905. doi: 10.1128/JVI.78.20.10888-10905.2004. J Virol. 2004. PMID: 15452209 Free PMC article.
-
Heterogeneous Nuclear Ribonucleoprotein A1 (hnRNP A1) and hnRNP A2 Inhibit Splicing to Human Papillomavirus 16 Splice Site SA409 through a UAG-Containing Sequence in the E7 Coding Region.J Virol. 2020 Sep 29;94(20):e01509-20. doi: 10.1128/JVI.01509-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32759322 Free PMC article.
-
Papillomavirus transcripts and posttranscriptional regulation.Virology. 2013 Oct;445(1-2):187-96. doi: 10.1016/j.virol.2013.04.034. Epub 2013 May 23. Virology. 2013. PMID: 23706315 Review.
-
Splicing and Polyadenylation of Human Papillomavirus Type 16 mRNAs.Int J Mol Sci. 2017 Feb 9;18(2):366. doi: 10.3390/ijms18020366. Int J Mol Sci. 2017. PMID: 28208770 Free PMC article. Review.
Cited by
-
The Role of RNA Splicing Factors in Cancer: Regulation of Viral and Human Gene Expression in Human Papillomavirus-Related Cervical Cancer.Front Cell Dev Biol. 2020 Jun 12;8:474. doi: 10.3389/fcell.2020.00474. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32596243 Free PMC article. Review.
-
HPV16 and HPV18 Genome Structure, Expression, and Post-Transcriptional Regulation.Int J Mol Sci. 2022 Apr 29;23(9):4943. doi: 10.3390/ijms23094943. Int J Mol Sci. 2022. PMID: 35563334 Free PMC article. Review.
-
Role of the DNA Damage Response in Human Papillomavirus RNA Splicing and Polyadenylation.Int J Mol Sci. 2018 Jun 12;19(6):1735. doi: 10.3390/ijms19061735. Int J Mol Sci. 2018. PMID: 29895741 Free PMC article. Review.
-
Overexpression of m6A-factors METTL3, ALKBH5, and YTHDC1 alters HPV16 mRNA splicing.Virus Genes. 2022 Apr;58(2):98-112. doi: 10.1007/s11262-022-01889-6. Epub 2022 Feb 21. Virus Genes. 2022. PMID: 35190939 Free PMC article.
-
Identification of heterogenous nuclear ribonucleoproteins (hnRNPs) and serine- and arginine-rich (SR) proteins that induce human papillomavirus type 16 late gene expression and alter L1 mRNA splicing.Arch Virol. 2022 Feb;167(2):563-570. doi: 10.1007/s00705-021-05317-2. Epub 2021 Dec 3. Arch Virol. 2022. PMID: 34860285 Free PMC article.
References
-
- Walboomers J. M., Jacobs M. V., Manos M. M., Bosch F. X., Kummer J. A., Shah K. V., Snijders P. J., Peto J., Meijer C. J., Muñoz N. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 - PubMed
-
- Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L., Cogliano V., and WHO International Agency for Research on Cancer Monograph Working Group (2009) A review of human carcinogens: part B: biological agents. Lancet Oncol. 10, 321–322 - PubMed
-
- zur Hausen H. (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2, 342–350 - PubMed
-
- Howley P. M., Lowy D. R. (2006) in Virology, 5th Ed (Knipe D. M., Howley P. M., eds) pp 2299–2354, Lippincott, Philadelphia
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

