DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity
- PMID: 25878287
- PMCID: PMC4397609
- DOI: 10.1523/JNEUROSCI.4571-14.2015
DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity
Abstract
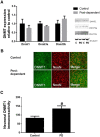
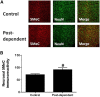
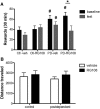
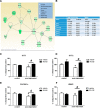
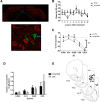
Recent studies have suggested an association between alcoholism and DNA methylation, a mechanism that can mediate long-lasting changes in gene transcription. Here, we examined the contribution of DNA methylation to the long-term behavioral and molecular changes induced by a history of alcohol dependence. In search of mechanisms underlying persistent rather than acute dependence-induced neuroadaptations, we studied the role of DNA methylation regulating medial prefrontal cortex (mPFC) gene expression and alcohol-related behaviors in rats 3 weeks into abstinence following alcohol dependence. Postdependent rats showed escalated alcohol intake, which was associated with increased DNA methylation as well as decreased expression of genes encoding synaptic proteins involved in neurotransmitter release in the mPFC. Infusion of the DNA methyltransferase inhibitor RG108 prevented both escalation of alcohol consumption and dependence-induced downregulation of 4 of the 7 transcripts modified in postdependent rats. Specifically, RG108 treatment directly reversed both downregulation of synaptotagmin 2 (Syt2) gene expression and hypermethylation on CpG#5 of its first exon. Lentiviral inhibition of Syt2 expression in the mPFC increased aversion-resistant alcohol drinking, supporting a mechanistic role of Syt2 in compulsive-like behavior. Our findings identified a functional role of DNA methylation in alcohol dependence-like behavioral phenotypes and a candidate gene network that may mediate its effects. Together, these data provide novel evidence for DNA methyltransferases as potential therapeutic targets in alcoholism.
Keywords: DNA methylation; DNMT; alcoholism; epigenetics; neurotransmitter release; plasticity.
Copyright © 2015 the authors 0270-6474/15/356153-12$15.00/0.
Figures

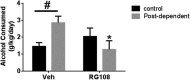

References
-
- Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
