Comparative Evaluation of Two PCR-Based Methods for Detection of Methicillin-Resistant Staphylococcus aureus (MRSA): Xpert MRSA Gen 3 and BD-Max MRSA XT
- PMID: 25878336
- PMCID: PMC4432069
- DOI: 10.1128/JCM.03679-14
Comparative Evaluation of Two PCR-Based Methods for Detection of Methicillin-Resistant Staphylococcus aureus (MRSA): Xpert MRSA Gen 3 and BD-Max MRSA XT
Abstract
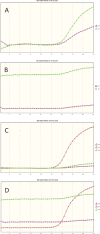
We compared two walk-away molecular diagnostic assays, the GeneXpert MRSA Gen 3 assay and the BD-Max MRSA XT assay. A total of 119 prospective swabs and 36 culture-positive samples were tested. Xpert MRSA Gen 3 had sensitivity of 95.7% and specificity of 100% versus 87.5% and 97.1% for BD-Max. The difference in agreement with the enriched culture results was significantly in favor of the Xpert assay (P < 0.02, McNemar nonparametric text).
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Performance of the Cepheid Xpert® SA Nasal Complete PCR assay compared to culture for detection of methicillin-sensitive and methicillin-resistant Staphylococcus aureus colonization.Diagn Microbiol Infect Dis. 2014 Sep;80(1):32-4. doi: 10.1016/j.diagmicrobio.2014.05.019. Epub 2014 May 21. Diagn Microbiol Infect Dis. 2014. PMID: 24952987
-
Detection of mecA- and mecC-Positive Methicillin-Resistant Staphylococcus aureus (MRSA) Isolates by the New Xpert MRSA Gen 3 PCR Assay.J Clin Microbiol. 2016 Jan;54(1):180-4. doi: 10.1128/JCM.02081-15. Epub 2015 Oct 21. J Clin Microbiol. 2016. PMID: 26491186 Free PMC article.
-
Comparison of the BD MAX MRSA XT to the Cepheid™ Xpert® MRSA assay for the molecular detection of methicillin-resistant Staphylococcus aureus from nasal swabs.Diagn Microbiol Infect Dis. 2017 Apr;87(4):308-310. doi: 10.1016/j.diagmicrobio.2016.12.011. Epub 2016 Dec 16. Diagn Microbiol Infect Dis. 2017. PMID: 28094151
-
The use of molecular methods for the detection and identification of methicillin-resistant Staphylococcus aureus.Biomark Med. 2014;8(9):1115-25. doi: 10.2217/bmm.14.60. Biomark Med. 2014. PMID: 25402581 Review.
-
[Clinical Significance of Next-Generation Molecular Testing Systems for the Diagnosis of Infectious Diseases and Infection Control].Rinsho Byori. 2014 Oct;62(10):1003-12. Rinsho Byori. 2014. PMID: 27526549 Review. Japanese.
Cited by
-
Characterization of SCCmec Instability in Methicillin-Resistant Staphylococcus aureus Affecting Adjacent Chromosomal Regions, Including the Gene for Staphylococcal Protein A (spa).Antimicrob Agents Chemother. 2022 Apr 19;66(4):e0237421. doi: 10.1128/aac.02374-21. Epub 2022 Mar 7. Antimicrob Agents Chemother. 2022. PMID: 35254090 Free PMC article.
-
Evolving Rapid Methicillin-resistant Staphylococcus aureus Detection: Cover All the Bases.J Glob Infect Dis. 2017 Jan-Mar;9(1):18-22. doi: 10.4103/0974-777X.199997. J Glob Infect Dis. 2017. PMID: 28250621 Free PMC article. Review.
-
A Novel Assay for Detection of Methicillin-Resistant Staphylococcus aureus Directly From Clinical Samples.Front Microbiol. 2020 Jun 18;11:1295. doi: 10.3389/fmicb.2020.01295. eCollection 2020. Front Microbiol. 2020. PMID: 32625187 Free PMC article.
-
Clinical Relevance of Xpert MRSA/SA in Guiding Therapeutic Decisions for Staphylococcal Infections: A Diagnostic Test Accuracy Analysis.Infect Dis Ther. 2022 Jun;11(3):1205-1227. doi: 10.1007/s40121-022-00632-w. Epub 2022 Apr 22. Infect Dis Ther. 2022. PMID: 35451743 Free PMC article.
-
Evaluation of BD Max StaphSR and BD Max MRSAXT Assays Using ESwab-Collected Specimens.J Clin Microbiol. 2015 Aug;53(8):2525-9. doi: 10.1128/JCM.00970-15. Epub 2015 May 27. J Clin Microbiol. 2015. PMID: 26019193 Free PMC article.
References
-
- Schweizer ML, Furuno JP, Harris AD, Johnson JK, Shardell MD, McGregor JC, Thom KA, Cosgrove SE, Sakoulas G, Perencevich EN. 2011. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis 11:279. doi:10.1186/1471-2334-11-279. - DOI - PMC - PubMed
-
- Stryjewski ME, Szczech LA, Benjamin DK Jr, Inrig JK, Kanafani ZA, Engemann JJ, Chu VH, Joyce MJ, Reller LB, Corey GR, Fowler VG Jr. 2007. Use of vancomycin or first-generation cephalosporins for the treatment of hemodialysis-dependent patients with methicillin-susceptible Staphylococcus aureus bacteremia. Clin Infect Dis 44:190–196. doi:10.1086/510386. - DOI - PubMed
-
- Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, van Belkum A, Ouellette M, Roy PH, Bergeron MG. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol 42:1875–1884. doi:10.1128/JCM.42.5.1875-1884.2004. - DOI - PMC - PubMed
-
- Trouillet-Assant S, Rasigade JP, Lustig S, Lhoste Y, Valour F, Guerin C, Aubrun F, Tigaud S, Laurent F. 2013. Ward-specific rates of nasal cocolonization with methicillin-susceptible and -resistant Staphylococcus spp. and potential impact on molecular methicillin-resistant Staphylococcus aureus screening tests. J Clin Microbiol 51:2418–2420. doi:10.1128/JCM.00491-13. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

