CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers
- PMID: 25879410
- PMCID: PMC4428112
- DOI: 10.1186/s12864-015-1419-2
CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers
Abstract
Background: The problem of supervised DNA sequence classification arises in several fields of computational molecular biology. Although this problem has been extensively studied, it is still computationally challenging due to size of the datasets that modern sequencing technologies can produce.
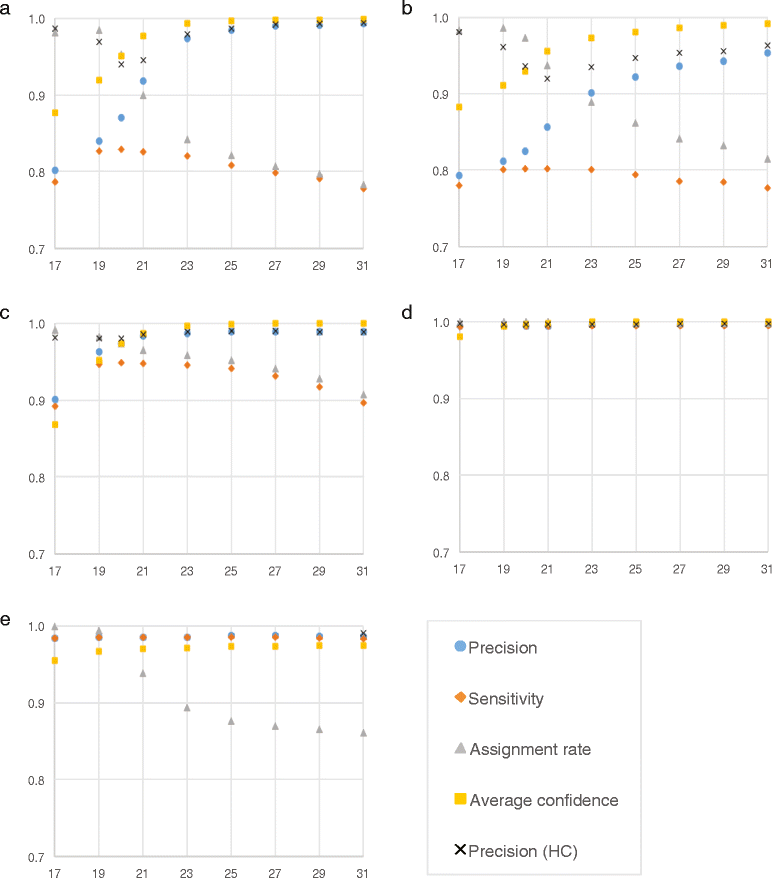
Results: We introduce CLARK a novel approach to classify metagenomic reads at the species or genus level with high accuracy and high speed. Extensive experimental results on various metagenomic samples show that the classification accuracy of CLARK is better or comparable to the best state-of-the-art tools and it is significantly faster than any of its competitors. In its fastest single-threaded mode CLARK classifies, with high accuracy, about 32 million metagenomic short reads per minute. CLARK can also classify BAC clones or transcripts to chromosome arms and centromeric regions.
Conclusions: CLARK is a versatile, fast and accurate sequence classification method, especially useful for metagenomics and genomics applications. It is freely available at http://clark.cs.ucr.edu/ .
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

