BRCA1 functions as a novel transcriptional cofactor in HIV-1 infection
- PMID: 25879655
- PMCID: PMC4359766
- DOI: 10.1186/s12985-015-0266-8
BRCA1 functions as a novel transcriptional cofactor in HIV-1 infection
Abstract
Background: Viruses have naturally evolved elegant strategies to manipulate the host's cellular machinery, including ways to hijack cellular DNA repair proteins to aid in their own replication. Retroviruses induce DNA damage through integration of their genome into host DNA. DNA damage signaling proteins including ATR, ATM and BRCA1 contribute to multiple steps in the HIV-1 life cycle, including integration and Vpr-induced G2/M arrest. However, there have been no studies to date regarding the role of BRCA1 in HIV-1 transcription.
Methods: Here we performed various transcriptional analyses to assess the role of BRCA1 in HIV-1 transcription by overexpression, selective depletion, and treatment with small molecule inhibitors. We examined association of Tat and BRCA1 through in vitro binding assays, as well as BRCA1-LTR association by chromatin immunoprecipitation.
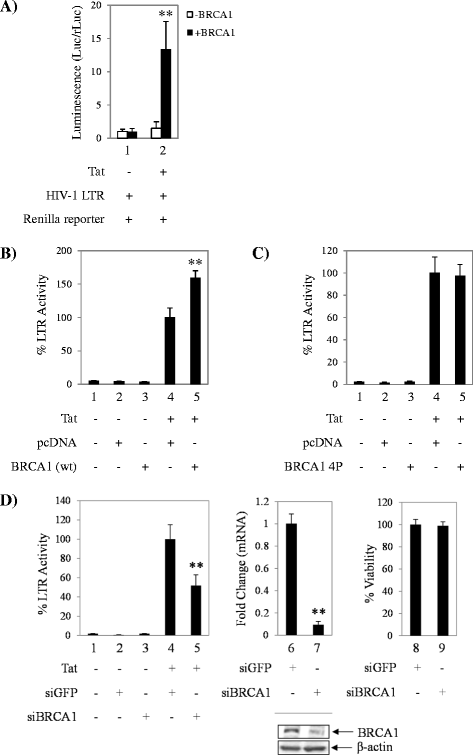
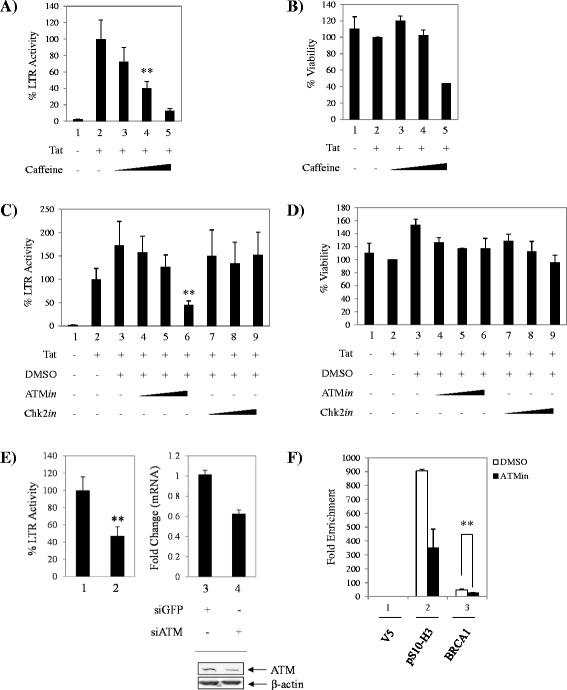
Results: BRCA1 was found to be important for viral transcription as cells that lack BRCA1 displayed severely reduced HIV-1 Tat-dependent transcription, and gain or loss-of-function studies resulted in enhanced or decreased transcription. Moreover, Tat was detected in complex with BRCA1 aa504-802. Small molecule inhibition of BRCA1 phosphorylation effector kinases, ATR and ATM, decreased Tat-dependent transcription, whereas a Chk2 inhibitor showed no effect. Furthermore, BRCA1 was found at the viral promoter and treatment with curcumin and ATM inhibitors decreased BRCA1 LTR occupancy. Importantly, these findings were validated in a highly relevant model of HIV infection and are indicative of BRCA1 phosphorylation affecting Tat-dependent transcription.
Conclusions: BRCA1 presence at the HIV-1 promoter highlights a novel function of the multifaceted protein in HIV-1 infection. The BRCA1 pathway or enzymes that phosphorylate BRCA1 could potentially be used as complementary host-based treatment for combined antiretroviral therapy, as there are multiple potent ATM inhibitors in development as chemotherapeutics.
Figures




Similar articles
-
ZBTB2 represses HIV-1 transcription and is regulated by HIV-1 Vpr and cellular DNA damage responses.PLoS Pathog. 2021 Feb 26;17(2):e1009364. doi: 10.1371/journal.ppat.1009364. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33635925 Free PMC article.
-
FoxO4 negatively controls Tat-mediated HIV-1 transcription through the post-transcriptional suppression of Tat encoding mRNA.J Gen Virol. 2017 Jul;98(7):1864-1878. doi: 10.1099/jgv.0.000837. Epub 2017 Jul 12. J Gen Virol. 2017. PMID: 28699853
-
HIV-1 Tat phosphorylation on Ser-16 residue modulates HIV-1 transcription.Retrovirology. 2018 May 23;15(1):39. doi: 10.1186/s12977-018-0422-5. Retrovirology. 2018. PMID: 29792216 Free PMC article.
-
Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription.Curr Pharm Des. 2017;23(28):4079-4090. doi: 10.2174/1381612823666170622104355. Curr Pharm Des. 2017. PMID: 28641539 Free PMC article. Review.
-
Tat is a multifunctional viral protein that modulates cellular gene expression and functions.Oncotarget. 2017 Apr 18;8(16):27569-27581. doi: 10.18632/oncotarget.15174. Oncotarget. 2017. PMID: 28187438 Free PMC article. Review.
Cited by
-
The consequences of viral infection on host DNA damage response: a focus on SARS-CoVs.J Genet Eng Biotechnol. 2022 Jul 13;20(1):104. doi: 10.1186/s43141-022-00388-3. J Genet Eng Biotechnol. 2022. PMID: 35829826 Free PMC article. Review.
-
Gene-expression reversal of lncRNAs and associated mRNAs expression in active vs latent HIV infection.Sci Rep. 2016 Oct 19;6:34862. doi: 10.1038/srep34862. Sci Rep. 2016. PMID: 27756902 Free PMC article.
-
Filgotinib suppresses HIV-1-driven gene transcription by inhibiting HIV-1 splicing and T cell activation.J Clin Invest. 2020 Sep 1;130(9):4969-4984. doi: 10.1172/JCI137371. J Clin Invest. 2020. PMID: 32573496 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

