New insights into the evolutionary history of plant sorbitol dehydrogenase
- PMID: 25879735
- PMCID: PMC4404067
- DOI: 10.1186/s12870-015-0478-5
New insights into the evolutionary history of plant sorbitol dehydrogenase
Abstract
Background: Sorbitol dehydrogenase (SDH, EC 1.1.1.14) is the key enzyme involved in sorbitol metabolism in higher plants. SDH genes in some Rosaceae species could be divided into two groups. L-idonate-5-dehydrogenase (LIDH, EC 1.1.1.264) is involved in tartaric acid (TA) synthesis in Vitis vinifera and is highly homologous to plant SDHs. Despite efforts to understand the biological functions of plant SDH, the evolutionary history of plant SDH genes and their phylogenetic relationship with the V. vinifera LIDH gene have not been characterized.
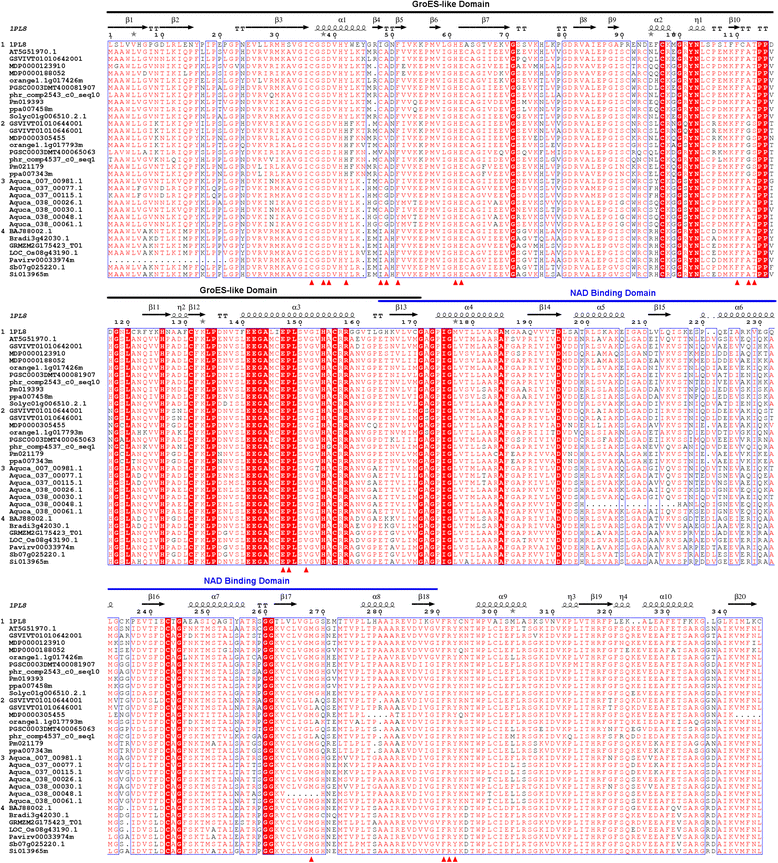
Results: A total of 92 SDH genes were identified from 42 angiosperm species. SDH genes have been highly duplicated within the Rosaceae family while monocot, Brassicaceae and most Asterid species exhibit singleton SDH genes. Core Eudicot SDHs have diverged into two phylogenetic lineages, now classified as SDH Class I and SDH Class II. V. vinifera LIDH was identified as a Class II SDH. Tandem duplication played a dominant role in the expansion of plant SDH family and Class II SDH genes were positioned in tandem with Class I SDH genes in several plant genomes. Protein modelling analyses of V. vinifera SDHs revealed 19 putative active site residues, three of which exhibited amino acid substitutions between Class I and Class II SDHs and were influenced by positive natural selection in the SDH Class II lineage. Gene expression analyses also demonstrated a clear transcriptional divergence between Class I and Class II SDH genes in V. vinifera and Citrus sinensis (orange).
Conclusions: Phylogenetic, natural selection and synteny analyses provided strong support for the emergence of SDH Class II by positive natural selection after tandem duplication in the common ancestor of core Eudicot plants. The substitutions of three putative active site residues might be responsible for the unique enzyme activity of V. vinifera LIDH, which belongs to SDH Class II and represents a novel function of SDH in V. vinifera that may be true also of other Class II SDHs. Gene expression analyses also supported the divergence of SDH Class II at the expression level. This study will facilitate future research into understanding the biological functions of plant SDHs.
Figures





References
-
- Iwata T, Hoog JO, Reddy VN, Carper D. Cloning of the human Sorbitol Dehydrogenase gene. Invest Ophth Vis Sci. 1993;34(4):712–2.
-
- Karlsson C, Jornvall H, Hoog JO. Sorbitol Dehydrogenase - cDNA coding for the rat enzyme - variations within the Alcohol-Dehydrogenase family independent of quaternary structure and metal content. Eur J Biochem. 1991;198(3):761–5. - PubMed
-
- Wang T, Hou M, Zhao N, Chen Y, Lv Y, Li Z, et al. Cloning and expression of the sorbitol dehydrogenase gene during embryonic development and temperature stress in Artemia sinica. Gene. 2013;521(2):296–302. - PubMed
-
- Niimi T, Yamashita O, Yaginuma T. A cold-inducible Bombyx gene encoding a protein similar to mammalian Sorbitol Dehydrogenase - yolk nuclei-dependent gene-expression in diapause eggs. Eur J Biochem. 1993;213(3):1125–31. - PubMed
-
- Sarthy AV, Schopp C, Idler KB. Cloning and sequence determination of the gene encoding Sorbitol Dehydrogenase from Saccharomyces cerevisiae. Gene. 1994;140(1):121–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

