The emerging landscape of small nucleolar RNAs in cell biology
- PMID: 25879954
- PMCID: PMC4696412
- DOI: 10.1002/wrna.1284
The emerging landscape of small nucleolar RNAs in cell biology
Abstract
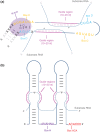
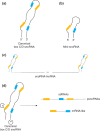
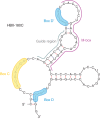
Small nucleolar RNAs (snoRNAs) are a large class of small noncoding RNAs present in all eukaryotes sequenced thus far. As a family, they have been well characterized as playing a central role in ribosome biogenesis, guiding either the sequence-specific chemical modification of pre-rRNA (ribosomal RNA) or its processing. However, in higher eukaryotes, numerous orphan snoRNAs were described over a decade ago, with no known target or ascribed function, suggesting the possibility of alternative cellular functionality. In recent years, thanks in great part to advances in sequencing methodologies, we have seen many examples of the diversity that exists in the snoRNA family on multiple levels. In this review, we discuss the identification of novel snoRNA members, of unexpected binding partners, as well as the clarification and extension of the snoRNA target space and the characterization of diverse new noncanonical functions, painting a new and extended picture of the snoRNA landscape. Under the deluge of novel features and functions that have recently come to light, snoRNAs emerge as a central, dynamic, and highly versatile group of small regulatory RNAs.
© 2015 The Authors. WIREs RNA published by John Wiley & Sons, Ltd.
Figures
References
-
- Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. - PubMed
-
- Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. - PubMed
-
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. - PubMed
-
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

