A pseudogene-signature in glioma predicts survival
- PMID: 25880120
- PMCID: PMC4364677
- DOI: 10.1186/s13046-015-0137-6
A pseudogene-signature in glioma predicts survival
Abstract
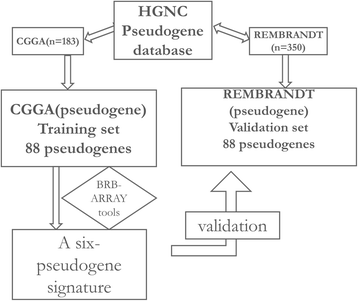
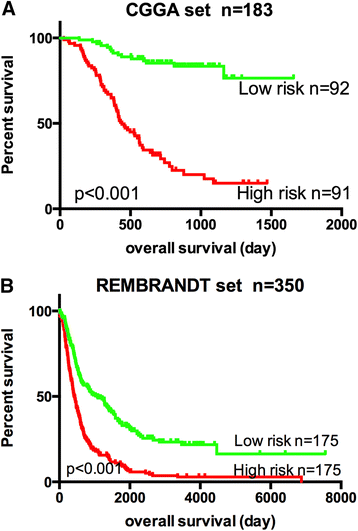
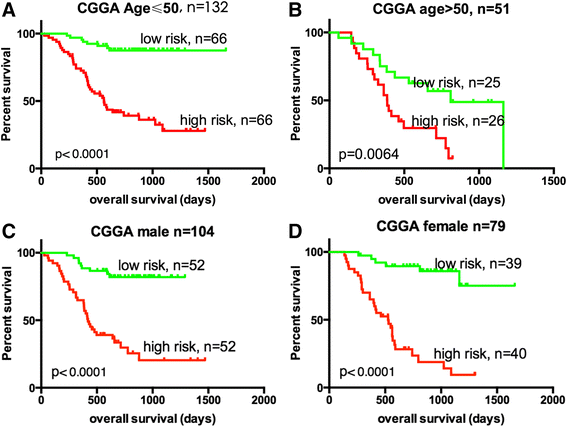
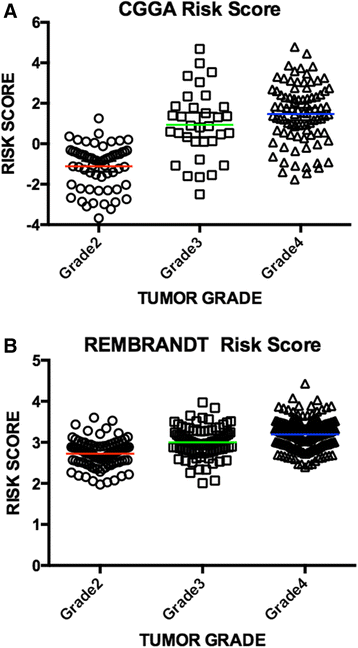
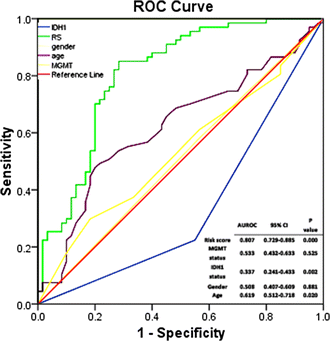
Pseudogene was recognized as a potential tumor suppressor or oncogene in varies of diseases, however its roles in glioma have not been investigated. Our study was to identify the pseudogene-signature that predicted glioma survival. Using a pseudogene-mining approach, we performed pseudogene expression profiling in 183 glioma samples from the Chinese Glioma Genome Atlas (CGGA) and set it as the training set. We found a six-pseudogene signature correlated with patients' clinical outcome via bioinformatics analyses (P ≤ 0.01), and validated it in the Repository of Molecular Brain Neoplasia Data (REMBRANDT) containing 350 cases. A formula calculating the risk score based on the six-pseudogene signature was introduced and the patients of CGGA set were classified into high-risk group and low-risk group with remarkably different survival (P < 0.001) based on their scores. The prognostic value of the signature was confirmed in the REMBRANDT set. Though the function of these pseudogenes is not clear, the identification of the prognostic pseudogenes indicated the potential roles of pseudogenes in glioma pathogenesis and they may have clinical implications in treating glioma.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

