Hyperphosphorylation of DegU cancels CcpA-dependent catabolite repression of rocG in Bacillus subtilis
- PMID: 25880922
- PMCID: PMC4348106
- DOI: 10.1186/s12866-015-0373-0
Hyperphosphorylation of DegU cancels CcpA-dependent catabolite repression of rocG in Bacillus subtilis
Abstract
Background: The two-component regulatory system, involving the histidine sensor kinase DegS and response regulator DegU, plays an important role to control various cell processes in the transition phase of Bacillus subtilis. The degU32 allele in strain 1A95 is characterized by the accumulation of phosphorylated form of DegU (DegU-P).
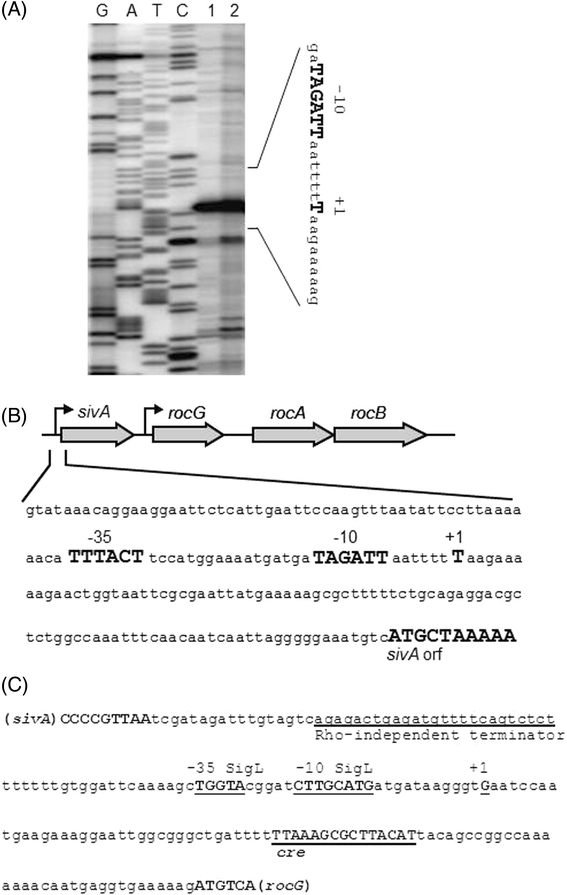
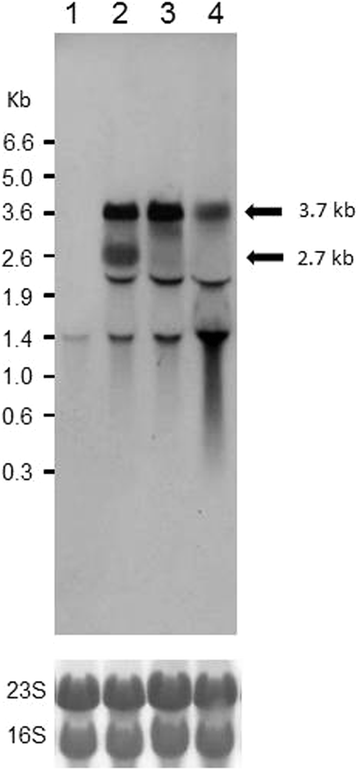
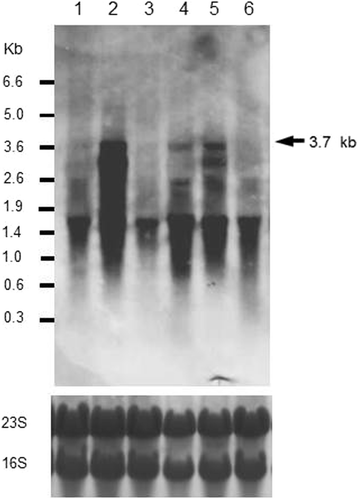
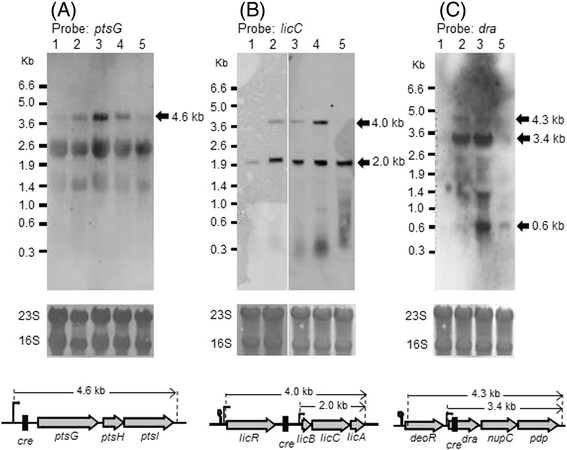
Results: Growing 1A95 cells elevated the pH of soytone-based medium more than the parental strain 168 after the onset of the transition phase. The rocG gene encodes a catabolic glutamate dehydrogenase that catalyzes one of the main ammonia-releasing reactions. Inactivation of rocG abolished 1A95-mediated increases in the pH of growth media. Thus, transcription of the rocG locus was examined, and a novel 3.7-kb transcript covering sivA, rocG, and rocA was found in 1A95 but not 168 cells. Increased intracellular fructose 1,6-bisphosphate (FBP) levels are known to activate the HPr kinase HPrK, and to induce formation of the P-Ser-HPr/CcpA complex, which binds to catabolite responsive elements (cre) and exerts CcpA-dependent catabolite repression. A putative cre found within the intergenic region between sivA and rocG, and inactivation of ccpA led to creation of the 3.7-kb transcript in 168 cells. Analyses of intermediates in central carbon metabolism revealed that intracellular FBP levels were lowered earlier in 1A95 than in 168 cells. A genome wide transcriptome analysis comparing 1A95 and 168 cells suggested similar events occurring in other catabolite repressive loci involving induction of lctE encoding lactate dehydrogenase.
Conclusions: Under physiological conditions the 3.7-kb rocG transcript may be tightly controlled by a roadblock mechanism involving P-Ser-HPr/CcpA in 168 cells, while in 1A95 cells abolished repression of the 3.7-kb transcript. Accumulation of DegU-P in 1A95 affects central carbon metabolism involving lctE enhanced by unknown mechanisms, downregulates FBP levels earlier, and inactivates HPrK to allow the 3.7-kb transcription, and thus similar events may occur in other catabolite repressive loci.
Figures




Similar articles
-
Regulated expression of HPrK/P does not affect carbon catabolite repression of the xyn operon and of rocG in Bacillus subtilis.FEMS Microbiol Lett. 2006 Jun;259(1):147-52. doi: 10.1111/j.1574-6968.2006.00260.x. FEMS Microbiol Lett. 2006. PMID: 16684115
-
Phosphorylation of either crh or HPr mediates binding of CcpA to the bacillus subtilis xyn cre and catabolite repression of the xyn operon.J Mol Biol. 1999 Feb 19;286(2):307-14. doi: 10.1006/jmbi.1998.2492. J Mol Biol. 1999. PMID: 9973552
-
Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression.Mol Microbiol. 2002 Feb;43(4):1039-52. doi: 10.1046/j.1365-2958.2002.02800.x. Mol Microbiol. 2002. PMID: 11929549
-
Carbon catabolite control of the metabolic network in Bacillus subtilis.Biosci Biotechnol Biochem. 2009 Feb;73(2):245-59. doi: 10.1271/bbb.80479. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202299 Review.
-
Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus.J Mol Microbiol Biotechnol. 2002 May;4(3):309-14. J Mol Microbiol Biotechnol. 2002. PMID: 11931563 Review.
Cited by
-
Secretion of the cytoplasmic and high molecular weight β-galactosidase of Paenibacillus wynnii with Bacillus subtilis.Microb Cell Fact. 2024 Jun 12;23(1):170. doi: 10.1186/s12934-024-02445-7. Microb Cell Fact. 2024. PMID: 38867249 Free PMC article.
-
DegU-mediated suppression of carbohydrate uptake in Listeria monocytogenes increases adaptation to oxidative stress.Appl Environ Microbiol. 2023 Oct 31;89(10):e0101723. doi: 10.1128/aem.01017-23. Epub 2023 Oct 3. Appl Environ Microbiol. 2023. PMID: 37787570 Free PMC article.
-
ComX-Induced Exoproteases Degrade ComX in Bacillus subtilis PS-216.Front Microbiol. 2018 Feb 1;9:105. doi: 10.3389/fmicb.2018.00105. eCollection 2018. Front Microbiol. 2018. PMID: 29449835 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

