BioPreDyn-bench: a suite of benchmark problems for dynamic modelling in systems biology
- PMID: 25880925
- PMCID: PMC4342829
- DOI: 10.1186/s12918-015-0144-4
BioPreDyn-bench: a suite of benchmark problems for dynamic modelling in systems biology
Abstract
Background: Dynamic modelling is one of the cornerstones of systems biology. Many research efforts are currently being invested in the development and exploitation of large-scale kinetic models. The associated problems of parameter estimation (model calibration) and optimal experimental design are particularly challenging. The community has already developed many methods and software packages which aim to facilitate these tasks. However, there is a lack of suitable benchmark problems which allow a fair and systematic evaluation and comparison of these contributions.
Results: Here we present BioPreDyn-bench, a set of challenging parameter estimation problems which aspire to serve as reference test cases in this area. This set comprises six problems including medium and large-scale kinetic models of the bacterium E. coli, baker's yeast S. cerevisiae, the vinegar fly D. melanogaster, Chinese Hamster Ovary cells, and a generic signal transduction network. The level of description includes metabolism, transcription, signal transduction, and development. For each problem we provide (i) a basic description and formulation, (ii) implementations ready-to-run in several formats, (iii) computational results obtained with specific solvers, (iv) a basic analysis and interpretation.
Conclusions: This suite of benchmark problems can be readily used to evaluate and compare parameter estimation methods. Further, it can also be used to build test problems for sensitivity and identifiability analysis, model reduction and optimal experimental design methods. The suite, including codes and documentation, can be freely downloaded from the BioPreDyn-bench website, https://sites.google.com/site/biopredynbenchmarks/ .
Figures
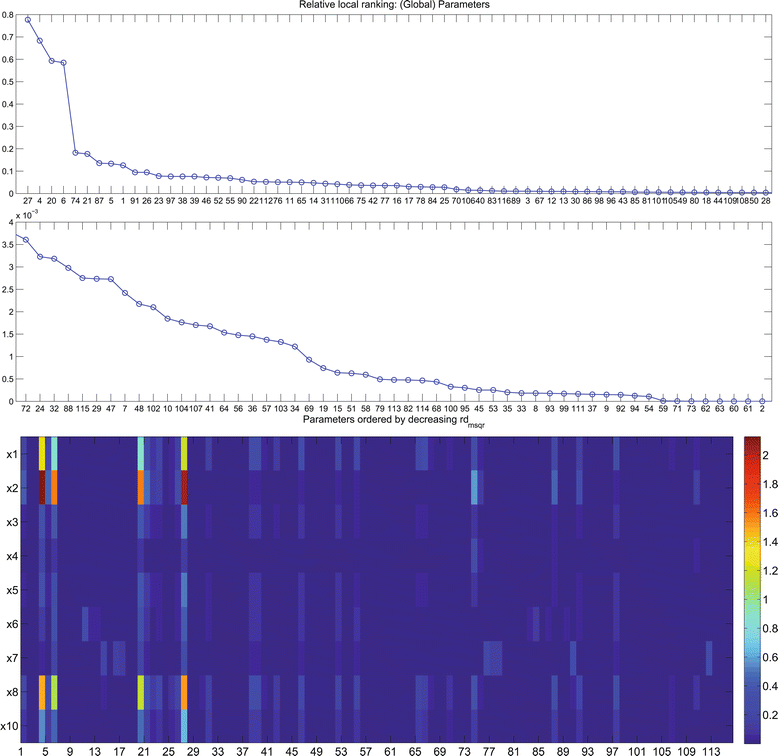



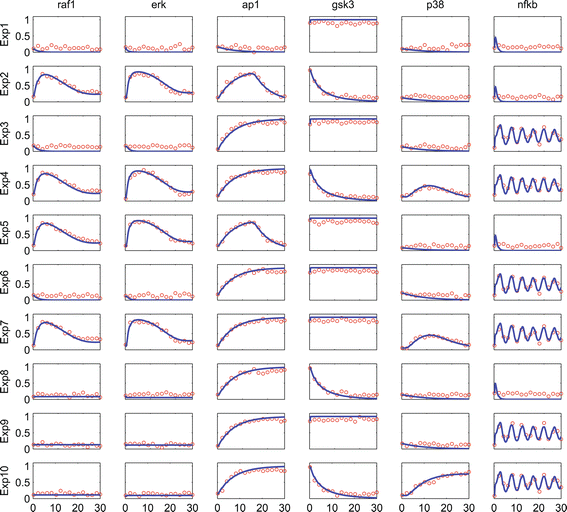
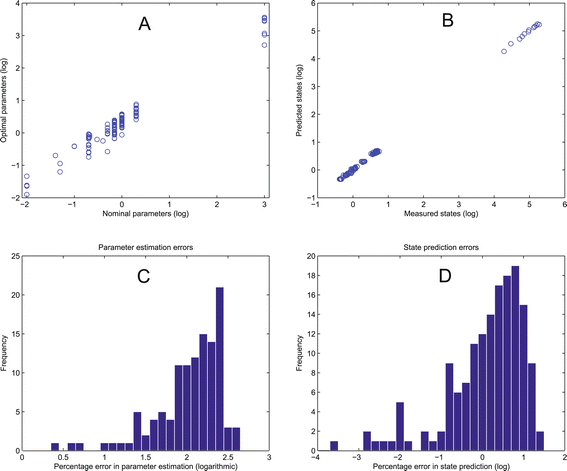
References
-
- Song H-S, DeVilbiss F, Ramkrishna D. Modeling metabolic systems: the need for dynamics. Curr Opin Chem Eng. 2013;2(4):373–82. doi: 10.1016/j.coche.2013.08.004. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

