A Point Mutation in the Ubiquitin Ligase RNF170 That Causes Autosomal Dominant Sensory Ataxia Destabilizes the Protein and Impairs Inositol 1,4,5-Trisphosphate Receptor-mediated Ca2+ Signaling
- PMID: 25882839
- PMCID: PMC4447968
- DOI: 10.1074/jbc.M115.655043
A Point Mutation in the Ubiquitin Ligase RNF170 That Causes Autosomal Dominant Sensory Ataxia Destabilizes the Protein and Impairs Inositol 1,4,5-Trisphosphate Receptor-mediated Ca2+ Signaling
Abstract
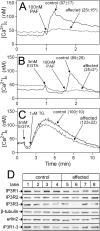
RNF170 is an endoplasmic reticulum membrane ubiquitin ligase that contributes to the ubiquitination of activated inositol 1,4,5-trisphosphate (IP3) receptors, and also, when point mutated (arginine to cysteine at position 199), causes autosomal dominant sensory ataxia (ADSA), a disease characterized by neurodegeneration in the posterior columns of the spinal cord. Here we demonstrate that this point mutation inhibits RNF170 expression and signaling via IP3 receptors. Inhibited expression of mutant RNF170 was seen in cells expressing exogenous RNF170 constructs and in ADSA lymphoblasts, and appears to result from enhanced RNF170 autoubiquitination and proteasomal degradation. The basis for these effects was probed via additional point mutations, revealing that ionic interactions between charged residues in the transmembrane domains of RNF170 are required for protein stability. In ADSA lymphoblasts, platelet-activating factor-induced Ca(2+) mobilization was significantly impaired, whereas neither Ca(2+) store content, IP3 receptor levels, nor IP3 production were altered, indicative of a functional defect at the IP3 receptor locus, which may be the cause of neurodegeneration. CRISPR/Cas9-mediated genetic deletion of RNF170 showed that RNF170 mediates the addition of all of the ubiquitin conjugates known to become attached to activated IP3 receptors (monoubiquitin and Lys(48)- and Lys(63)-linked ubiquitin chains), and that wild-type and mutant RNF170 have apparently identical ubiquitin ligase activities toward IP3 receptors. Thus, the Ca(2+) mobilization defect seen in ADSA lymphoblasts is apparently not due to aberrant IP3 receptor ubiquitination. Rather, the defect likely reflects abnormal ubiquitination of other substrates, or adaptation to the chronic reduction in RNF170 levels.
Keywords: E3 ubiquitin ligase; RNF170; calcium intracellular release; endoplasmic reticulum (ER); inositol trisphosphate receptor (InsP3R); neurodegenerative disease.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

