Assessing the impact of mutations found in next generation sequencing data over human signaling pathways
- PMID: 25883139
- PMCID: PMC4489259
- DOI: 10.1093/nar/gkv349
Assessing the impact of mutations found in next generation sequencing data over human signaling pathways
Abstract
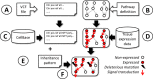
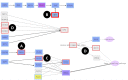
Modern sequencing technologies produce increasingly detailed data on genomic variation. However, conventional methods for relating either individual variants or mutated genes to phenotypes present known limitations given the complex, multigenic nature of many diseases or traits. Here we present PATHiVar, a web-based tool that integrates genomic variation data with gene expression tissue information. PATHiVar constitutes a new generation of genomic data analysis methods that allow studying variants found in next generation sequencing experiment in the context of signaling pathways. Simple Boolean models of pathways provide detailed descriptions of the impact of mutations in cell functionality so as, recurrences in functionality failures can easily be related to diseases, even if they are produced by mutations in different genes. Patterns of changes in signal transmission circuits, often unpredictable from individual genes mutated, correspond to patterns of affected functionalities that can be related to complex traits such as disease progression, drug response, etc. PATHiVar is available at: http://pathivar.babelomics.org.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Oti M., Brunner H.G. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. - PubMed
-
- Ioannidis J.P., Allison D.B., Ball C.A., Coulibaly I., Cui X., Culhane A.C., Falchi M., Furlanello C., Game L., Jurman G., et al. Repeatability of published microarray gene expression analyses. Nat. Genet. 2009;41:149–155. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

