Thioflavin T as a fluorescence probe for monitoring RNA metabolism at molecular and cellular levels
- PMID: 25883145
- PMCID: PMC4538803
- DOI: 10.1093/nar/gkv338
Thioflavin T as a fluorescence probe for monitoring RNA metabolism at molecular and cellular levels
Abstract
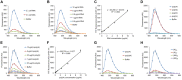
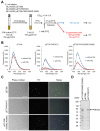
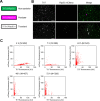
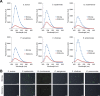
The intrinsically stochastic dynamics of mRNA metabolism have important consequences on gene regulation and non-genetic cell-to-cell variability; however, no generally applicable methods exist for studying such stochastic processes quantitatively. Here, we describe the use of the amyloid-binding probe Thioflavin T (ThT) for monitoring RNA metabolism in vitro and in vivo. ThT fluoresced strongly in complex with bacterial total RNA than with genomic DNA. ThT bound purine oligoribonucleotides preferentially over pyrimidine oligoribonucleotides and oligodeoxyribonucleotides. This property enabled quantitative real-time monitoring of poly(A) synthesis and phosphorolysis by polyribonucleotide phosphorylase in vitro. Cellular analyses, in combination with genetic approaches and the transcription-inhibitor rifampicin treatment, demonstrated that ThT mainly stained mRNA in actively dividing Escherichia coli cells. ThT also facilitated mRNA metabolism profiling at the single-cell level in diverse bacteria. Furthermore, ThT can also be used to visualise transitions between non-persister and persister cell states, a phenomenon of isogenic subpopulations of antibiotic-sensitive bacteria that acquire tolerance to multiple antibiotics due to stochastically induced dormant states. Collectively, these results suggest that probing mRNA dynamics with ThT is a broadly applicable approach ranging from the molecular level to the single-cell level.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. - PubMed
-
- Ozbudak E.M., Thattai M., Kurtser I., Grossman A.D., van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31:69–73. - PubMed
-
- Blake W.J., KAErn M., Cantor C.R., Collins J.J. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. - PubMed
-
- Kussell E., Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. - PubMed
-
- Süe G.M., Kulkarni R.P., Dworkin J., Garcia-Ojalvo J., Elowitz M.B. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

