I-TASSER server: new development for protein structure and function predictions
- PMID: 25883148
- PMCID: PMC4489253
- DOI: 10.1093/nar/gkv342
I-TASSER server: new development for protein structure and function predictions
Abstract
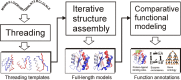
The I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER) is an online resource for automated protein structure prediction and structure-based function annotation. In I-TASSER, structural templates are first recognized from the PDB using multiple threading alignment approaches. Full-length structure models are then constructed by iterative fragment assembly simulations. The functional insights are finally derived by matching the predicted structure models with known proteins in the function databases. Although the server has been widely used for various biological and biomedical investigations, numerous comments and suggestions have been reported from the user community. In this article, we summarize recent developments on the I-TASSER server, which were designed to address the requirements from the user community and to increase the accuracy of modeling predictions. Focuses have been made on the introduction of new methods for atomic-level structure refinement, local structure quality estimation and biological function annotations. We expect that these new developments will improve the quality of the I-TASSER server and further facilitate its use by the community for high-resolution structure and function prediction.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
I-TASSER: a unified platform for automated protein structure and function prediction.Nat Protoc. 2010 Apr;5(4):725-38. doi: 10.1038/nprot.2010.5. Epub 2010 Mar 25. Nat Protoc. 2010. PMID: 20360767 Free PMC article.
-
I-TASSER-MR: automated molecular replacement for distant-homology proteins using iterative fragment assembly and progressive sequence truncation.Nucleic Acids Res. 2017 Jul 3;45(W1):W429-W434. doi: 10.1093/nar/gkx349. Nucleic Acids Res. 2017. PMID: 28472524 Free PMC article.
-
LOMETS2: improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins.Nucleic Acids Res. 2019 Jul 2;47(W1):W429-W436. doi: 10.1093/nar/gkz384. Nucleic Acids Res. 2019. PMID: 31081035 Free PMC article.
-
Computational tools for protein modeling.Curr Protein Pept Sci. 2000 Jul;1(1):1-21. doi: 10.2174/1389203003381469. Curr Protein Pept Sci. 2000. PMID: 12369918 Review.
-
I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction.Nat Protoc. 2022 Oct;17(10):2326-2353. doi: 10.1038/s41596-022-00728-0. Epub 2022 Aug 5. Nat Protoc. 2022. PMID: 35931779 Review.
Cited by
-
Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) lncRNA Conformational Dynamics in Complex with RNA-Binding Protein with Serine-Rich Domain 1 (RNPS1) in the Pan-cancer Splicing and Gene Expression.ACS Omega. 2024 Oct 3;9(41):42212-42226. doi: 10.1021/acsomega.4c04467. eCollection 2024 Oct 15. ACS Omega. 2024. PMID: 39431102 Free PMC article.
-
Co-Regulation Mechanism of Host p53 and Fos in Transcriptional Activation of ILTV Immediate-Early Gene ICP4.Microorganisms. 2024 Oct 16;12(10):2069. doi: 10.3390/microorganisms12102069. Microorganisms. 2024. PMID: 39458378 Free PMC article.
-
Structure/Function Analysis of Recurrent Mutations in SETD2 Protein Reveals a Critical and Conserved Role for a SET Domain Residue in Maintaining Protein Stability and Histone H3 Lys-36 Trimethylation.J Biol Chem. 2016 Sep 30;291(40):21283-21295. doi: 10.1074/jbc.M116.739375. Epub 2016 Aug 15. J Biol Chem. 2016. PMID: 27528607 Free PMC article.
-
Structural insights into membrane remodeling by SNX1.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2022614118. doi: 10.1073/pnas.2022614118. Proc Natl Acad Sci U S A. 2021. PMID: 33658379 Free PMC article.
-
Gene variants of coagulation related proteins that interact with SARS-CoV-2.PLoS Comput Biol. 2021 Mar 17;17(3):e1008805. doi: 10.1371/journal.pcbi.1008805. eCollection 2021 Mar. PLoS Comput Biol. 2021. PMID: 33730015 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

