Fully automated pipeline for detection of sex linked genes using RNA-Seq data
- PMID: 25884927
- PMCID: PMC4367819
- DOI: 10.1186/s12859-015-0509-0
Fully automated pipeline for detection of sex linked genes using RNA-Seq data
Abstract
Background: Sex chromosomes present a genomic region which to some extent, differs between the genders of a single species. Reliable high-throughput methods for detection of sex chromosomes specific markers are needed, especially in species where genome information is limited. Next generation sequencing (NGS) opens the door for identification of unique sequences or searching for nucleotide polymorphisms between datasets. A combination of classical genetic segregation analysis along with RNA-Seq data can present an ideal tool to map and identify sex chromosome-specific expressed markers. To address this challenge, we established genetic cross of dioecious plant Rumex acetosa and generated RNA-Seq data from both parental generation and male and female offspring.
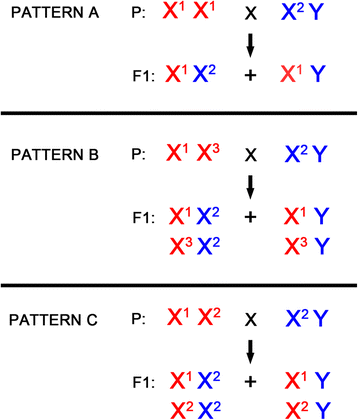
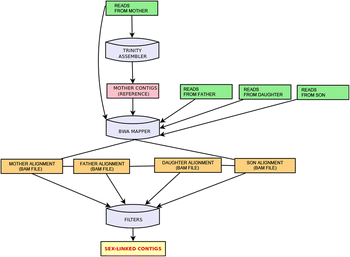
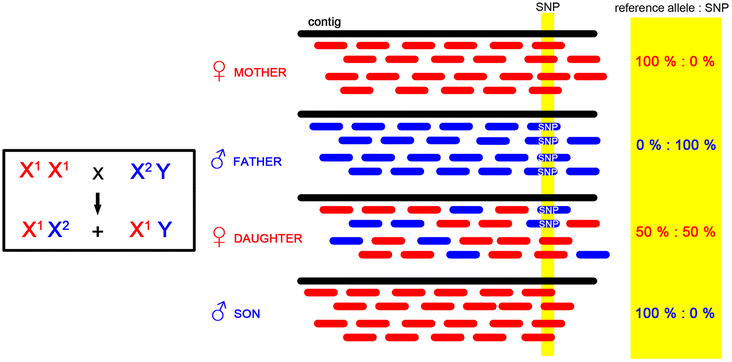
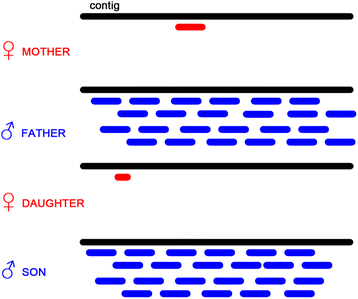
Results: We present a pipeline for detection of sex linked genes based on nucleotide polymorphism analysis. In our approach, tracking of nucleotide polymorphisms is carried out using a cross of preferably distant populations. For this reason, only 4 datasets are needed - reads from high-throughput sequencing platforms for parent generation (mother and father) and F1 generation (male and female progeny). Our pipeline uses custom scripts together with external assembly, mapping and variant calling software. Given the resource-intensive nature of the computation, servers with high capacity are a requirement. Therefore, in order to keep this pipeline easily accessible and reproducible, we implemented it in Galaxy - an open, web-based platform for data-intensive biomedical research. Our tools are present in the Galaxy Tool Shed, from which they can be installed to any local Galaxy instance. As an output of the pipeline, user gets a FASTA file with candidate transcriptionally active sex-linked genes, sorted by their relevance. At the same time, a BAM file with identified genes and alignment of reads is also provided. Thus, polymorphisms following segregation pattern can be easily visualized, which significantly enhances primer design and subsequent steps of wet-lab verification.
Conclusions: Our pipeline presents a simple and freely accessible software tool for identification of sex chromosome linked genes in species without an existing reference genome. Based on combination of genetic crosses and RNA-Seq data, we have designed a high-throughput, cost-effective approach for a broad community of scientists focused on sex chromosome structure and evolution.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

