Natural disease history and characterisation of SUMF1 molecular defects in ten unrelated patients with multiple sulfatase deficiency
- PMID: 25885655
- PMCID: PMC4375846
- DOI: 10.1186/s13023-015-0244-7
Natural disease history and characterisation of SUMF1 molecular defects in ten unrelated patients with multiple sulfatase deficiency
Abstract
Background: Multiple sulfatase deficiency is a rare inherited metabolic disorder caused by mutations in the SUMF1 gene. The disease remains poorly known, often leading to a late diagnosis. This study aimed to provide improved knowledge of the disease, through complete clinical, biochemical, and molecular descriptions of a cohort of unrelated patients. The main objective was to identify prognostic markers, both phenotypic and genotypic, to accelerate the diagnosis and improve patient care.
Methods: The phenotypes of ten unrelated patients were fully documented at the clinical and biochemical levels. The long-term follow-up of each patient allowed correlations of the phenotypes to the disease outcomes. Each patient's molecular defects were also identified. Site-directed mutagenesis was used to individually express the mutants and assess their stability. Characterisation of the protein mutants was completed by in silico analyses based on sequence comparisons and structural models.
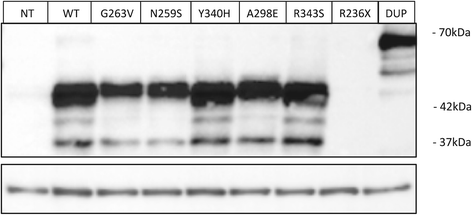
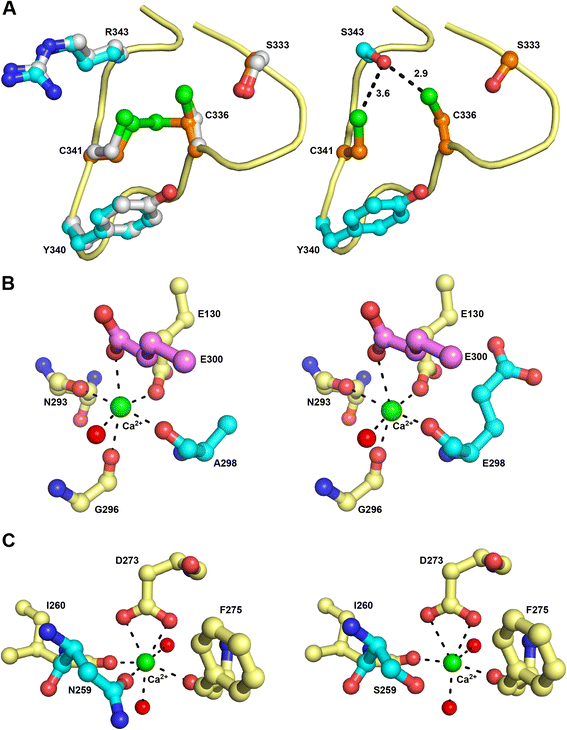
Results: The most severe cases were characterised by the presence of non-neurological symptoms as well as the occurrence of psychomotor regression before 2 years of age. Nine novel SUMF1 mutations were identified. Clinically severe forms were often associated with SUMF1 mutations that strongly affected the protein stability and/or catalytic function as predicted from in silico and western blot analyses.
Conclusions: This detailed clinical description and follow-up of a cohort of patients, together with the molecular characterisation of their underlying defects, contribute to improved knowledge of multiple sulfatase deficiency. Predictors of a bad prognosis were the presence of several non-neurological symptoms and the onset of psychomotor regression before 2 years of age. No strict correlation existed between in vitro residual sulfatase activity and disease severity. Genotype-phenotype correlations related to previously reported mutants were strengthened. These and previous observations allow not only improved prediction of the disease outcome but also provision of appropriate care for patients, in the expectation of specific treatment development.
Figures

Similar articles
-
[Clinical characterization and mutation identification for multiple sulfatase deficiency patients in China].Zhonghua Er Ke Za Zhi. 2013 Nov;51(11):836-41. Zhonghua Er Ke Za Zhi. 2013. PMID: 24484558 Chinese.
-
Molecular analysis of SUMF1 mutations: stability and residual activity of mutant formylglycine-generating enzyme determine disease severity in multiple sulfatase deficiency.Hum Mutat. 2008 Jan;29(1):205. doi: 10.1002/humu.9515. Hum Mutat. 2008. PMID: 18157819
-
Multiple sulfatase deficiency is due to hypomorphic mutations of the SUMF1 gene.Hum Mutat. 2007 Sep;28(9):928. doi: 10.1002/humu.9504. Hum Mutat. 2007. PMID: 17657823
-
Neonatal multiple sulfatase deficiency with a novel mutation and review of the literature.Turk J Pediatr. 2014 Jul-Aug;56(4):418-22. Turk J Pediatr. 2014. PMID: 25818962 Review.
-
Multiple Sulfatase Deficiency: A Disease Comprising Mucopolysaccharidosis, Sphingolipidosis, and More Caused by a Defect in Posttranslational Modification.Int J Mol Sci. 2020 May 13;21(10):3448. doi: 10.3390/ijms21103448. Int J Mol Sci. 2020. PMID: 32414121 Free PMC article. Review.
Cited by
-
Severe neonatal multiple sulfatase deficiency presenting with hydrops fetalis in a preterm birth patient.JIMD Rep. 2019 Aug 20;49(1):48-52. doi: 10.1002/jmd2.12074. eCollection 2019 Sep. JIMD Rep. 2019. PMID: 31497481 Free PMC article.
-
A homozygous missense variant of SUMF1 in the Bedouin population extends the clinical spectrum in ultrarare neonatal multiple sulfatase deficiency.Mol Genet Genomic Med. 2020 Sep;8(9):e1167. doi: 10.1002/mgg3.1167. Epub 2020 Feb 12. Mol Genet Genomic Med. 2020. PMID: 32048457 Free PMC article.
-
Boolean Modeling of Biological Network Applied to Protein-Protein Interaction Network of Autism Patients.Biology (Basel). 2024 Aug 10;13(8):606. doi: 10.3390/biology13080606. Biology (Basel). 2024. PMID: 39194544 Free PMC article.
-
Lysosomal Leukodystrophies Lysosomal Storage Diseases Associated With White Matter Abnormalities.J Child Neurol. 2019 May;34(6):339-358. doi: 10.1177/0883073819828587. Epub 2019 Feb 13. J Child Neurol. 2019. PMID: 30757954 Free PMC article. Review.
-
Severe central nervous system demyelination in Sanfilippo disease.Front Mol Neurosci. 2023 Dec 13;16:1323449. doi: 10.3389/fnmol.2023.1323449. eCollection 2023. Front Mol Neurosci. 2023. PMID: 38163061 Free PMC article.
References
-
- Austin J, McAfee D, Armstrong D, O’Rourke M, Shearer L, Bachhawat B. Abnormal sulphatase activities in two human diseases (metachromatic leucodystrophy and gargoylism) Biochem J. 1964;93:15C–17C. - PubMed
-
- Austin J, McAfee D, Armstrong D, O’Rourke M, Shearer L, Bachhawat B. Low sulfatase activities in metachromatic leukodystrophy (MLD). A controlled study of enzymes in 9 living and 4 autopsied patients with MLD. Trans Am Neurol Assoc. 1964;89:147–150. - PubMed
-
- Eto Y, Rampini S, Wiesmann U, Herschkowitz NN. Enzymic studies of sulphatases in tissues of the normal human and in metachromatic leukodystrophy with multiple sulphatase deficiencies: arylsulphatases A, B and C, cerebroside sulphatase, psychosine sulphatase and steroid sulphatases. J Neurochem. 1974;23:1161–1170. doi: 10.1111/j.1471-4159.1974.tb12213.x. - DOI - PubMed
-
- Eto Y, Gomibuchi I, Umezawa F, Tsuda T. Pathochemistry, pathogenesis and enzyme replacement in multiple-sulfatase deficiency. Enzyme. 1987;38:273–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

