MAGMA: generalized gene-set analysis of GWAS data
- PMID: 25885710
- PMCID: PMC4401657
- DOI: 10.1371/journal.pcbi.1004219
MAGMA: generalized gene-set analysis of GWAS data
Abstract
By aggregating data for complex traits in a biologically meaningful way, gene and gene-set analysis constitute a valuable addition to single-marker analysis. However, although various methods for gene and gene-set analysis currently exist, they generally suffer from a number of issues. Statistical power for most methods is strongly affected by linkage disequilibrium between markers, multi-marker associations are often hard to detect, and the reliance on permutation to compute p-values tends to make the analysis computationally very expensive. To address these issues we have developed MAGMA, a novel tool for gene and gene-set analysis. The gene analysis is based on a multiple regression model, to provide better statistical performance. The gene-set analysis is built as a separate layer around the gene analysis for additional flexibility. This gene-set analysis also uses a regression structure to allow generalization to analysis of continuous properties of genes and simultaneous analysis of multiple gene sets and other gene properties. Simulations and an analysis of Crohn's Disease data are used to evaluate the performance of MAGMA and to compare it to a number of other gene and gene-set analysis tools. The results show that MAGMA has significantly more power than other tools for both the gene and the gene-set analysis, identifying more genes and gene sets associated with Crohn's Disease while maintaining a correct type 1 error rate. Moreover, the MAGMA analysis of the Crohn's Disease data was found to be considerably faster as well.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
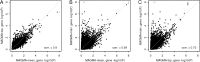
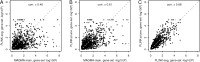
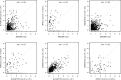
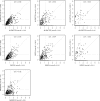
Similar articles
-
Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets.Genome Res. 2012 Feb;22(2):386-97. doi: 10.1101/gr.124370.111. Epub 2011 Sep 22. Genome Res. 2012. PMID: 21940837 Free PMC article.
-
FLAGS: A Flexible and Adaptive Association Test for Gene Sets Using Summary Statistics.Genetics. 2016 Mar;202(3):919-29. doi: 10.1534/genetics.115.185009. Epub 2016 Jan 15. Genetics. 2016. PMID: 26773050 Free PMC article.
-
How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures?Pac Symp Biocomput. 2018;23:228-239. Pac Symp Biocomput. 2018. PMID: 29218884 Free PMC article.
-
The statistical properties of gene-set analysis.Nat Rev Genet. 2016 Apr 12;17(6):353-64. doi: 10.1038/nrg.2016.29. Nat Rev Genet. 2016. PMID: 27070863 Review.
-
Annotating genetic variants to target genes using H-MAGMA.Nat Protoc. 2023 Jan;18(1):22-35. doi: 10.1038/s41596-022-00745-z. Epub 2022 Oct 26. Nat Protoc. 2023. PMID: 36289406 Free PMC article. Review.
Cited by
-
Single cell transcriptomes and multiscale networks from persons with and without Alzheimer's disease.Nat Commun. 2024 Jul 10;15(1):5815. doi: 10.1038/s41467-024-49790-0. Nat Commun. 2024. PMID: 38987616 Free PMC article.
-
Common risk variants in NPHS1 and TNFSF15 are associated with childhood steroid-sensitive nephrotic syndrome.Kidney Int. 2020 Nov;98(5):1308-1322. doi: 10.1016/j.kint.2020.05.029. Epub 2020 Jun 14. Kidney Int. 2020. PMID: 32554042 Free PMC article.
-
A Genome-Wide Association Study of Metabolic Syndrome in the Taiwanese Population.Nutrients. 2023 Dec 25;16(1):77. doi: 10.3390/nu16010077. Nutrients. 2023. PMID: 38201907 Free PMC article.
-
Independent replications and integrative analyses confirm TRANK1 as a susceptibility gene for bipolar disorder.Neuropsychopharmacology. 2021 May;46(6):1103-1112. doi: 10.1038/s41386-020-00788-4. Epub 2020 Aug 13. Neuropsychopharmacology. 2021. PMID: 32791513 Free PMC article.
-
The neocortical infrastructure for language involves region-specific patterns of laminar gene expression.Proc Natl Acad Sci U S A. 2024 Aug 20;121(34):e2401687121. doi: 10.1073/pnas.2401687121. Epub 2024 Aug 12. Proc Natl Acad Sci U S A. 2024. PMID: 39133845 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

