Transgenerational inheritance of diet-induced genome rearrangements in Drosophila
- PMID: 25885886
- PMCID: PMC4401788
- DOI: 10.1371/journal.pgen.1005148
Transgenerational inheritance of diet-induced genome rearrangements in Drosophila
Abstract
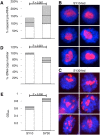
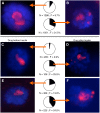
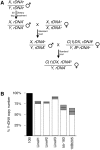
Ribosomal RNA gene (rDNA) copy number variation modulates heterochromatin formation and influences the expression of a large fraction of the Drosophila genome. This discovery, along with the link between rDNA, aging, and disease, high-lights the importance of understanding how natural rDNA copy number variation arises. Pursuing the relationship between rDNA expression and stability, we have discovered that increased dietary yeast concentration, emulating periods of dietary excess during life, results in somatic rDNA instability and copy number reduction. Modulation of Insulin/TOR signaling produces similar results, indicating a role for known nutrient sensing signaling pathways in this process. Furthermore, adults fed elevated dietary yeast concentrations produce offspring with fewer rDNA copies demonstrating that these effects also occur in the germline, and are transgenerationally heritable. This finding explains one source of natural rDNA copy number variation revealing a clear long-term consequence of diet.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

