DNA copy number analysis of metastatic urothelial carcinoma with comparison to primary tumors
- PMID: 25886454
- PMCID: PMC4392457
- DOI: 10.1186/s12885-015-1192-2
DNA copy number analysis of metastatic urothelial carcinoma with comparison to primary tumors
Abstract
Background: To date, there have been no reports characterizing the genome-wide somatic DNA chromosomal copy-number alteration landscape in metastatic urothelial carcinoma. We sought to characterize the DNA copy-number profile in a cohort of metastatic samples and compare them to a cohort of primary urothelial carcinoma samples in order to identify changes that are associated with progression from primary to metastatic disease.
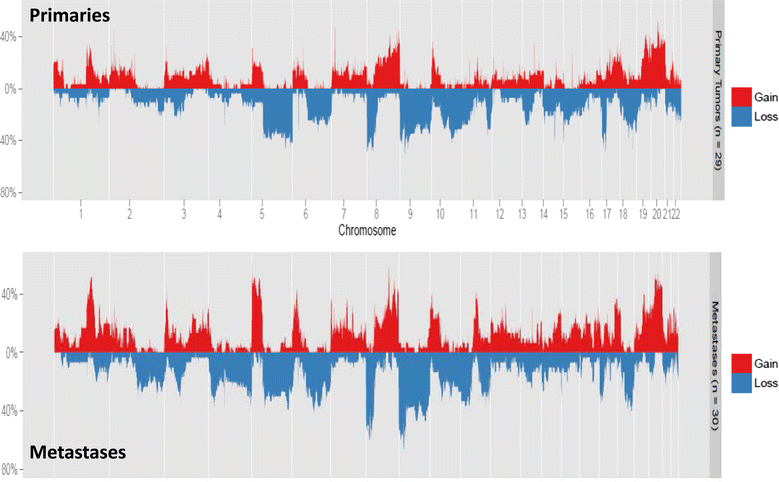
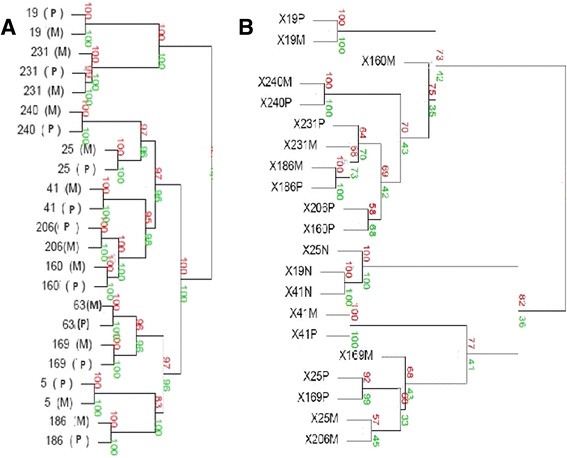
Methods: Using molecular inversion probe array analysis we compared genome-wide chromosomal copy-number alterations between 30 metastatic and 29 primary UC samples. Whole transcriptome RNA-Seq analysis was also performed in primary and matched metastatic samples which was available for 9 patients.
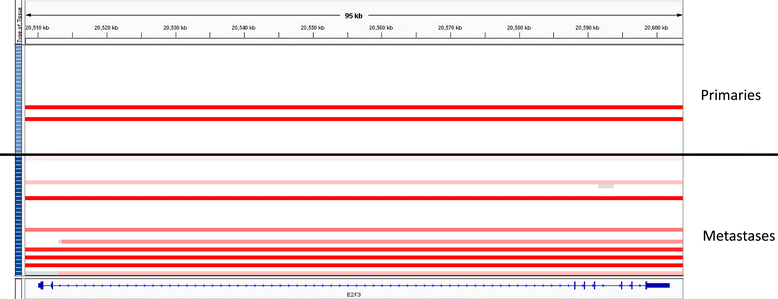
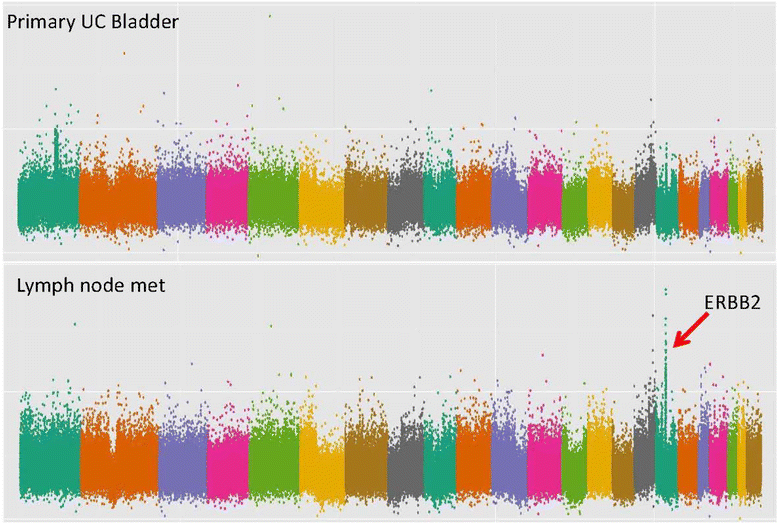
Results: Based on a focused analysis of 32 genes in which alterations may be clinically actionable, there were significantly more amplifications/deletions in metastases (8.6% vs 4.5%, p < 0.001). In particular, there was a higher frequency of E2F3 amplification in metastases (30% vs 7%, p = 0.046). Paired primary and metastatic tissue was available for 11 patients and 3 of these had amplifications of potential clinical relevance in metastases that were not in the primary tumor including ERBB2, CDK4, CCND1, E2F3, and AKT1. The transcriptional activity of these amplifications was supported by RNA expression data.
Conclusions: The discordance in alterations between primary and metastatic tissue may be of clinical relevance in the era of genomically directed precision cancer medicine.
Figures



References
-
- Eble JN, World Health Organization Classification of Tumours. Sauter G, Epstein JI, Sesterhenn IA. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyon, France: IARC Press; 2004.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

