Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin
- PMID: 25886480
- PMCID: PMC4423110
- DOI: 10.1186/s13059-015-0644-y
Age and sun exposure-related widespread genomic blocks of hypomethylation in nonmalignant skin
Abstract
Background: Aging and sun exposure are the leading causes of skin cancer. It has been shown that epigenetic changes, such as DNA methylation, are well established mechanisms for cancer, and also have emerging roles in aging and common disease. Here, we directly ask whether DNA methylation is altered following skin aging and/or chronic sun exposure in humans.
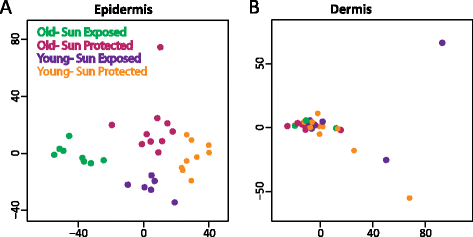
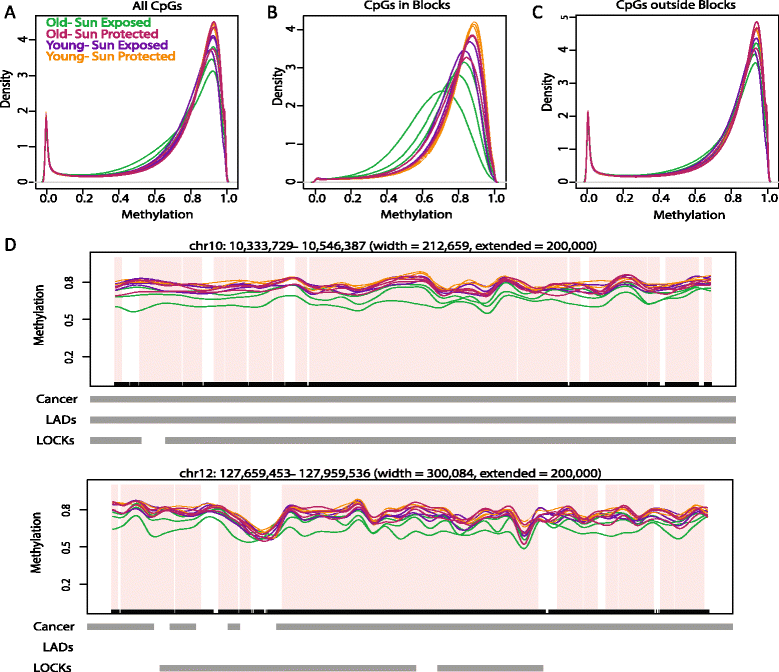
Results: We compare epidermis and dermis of both sun-protected and sun-exposed skin derived from younger subjects (under 35 years old) and older subjects (over 60 years old), using the Infinium HumanMethylation450 array and whole genome bisulfite sequencing. We observe large blocks of the genome that are hypomethylated in older, sun-exposed epidermal samples, with the degree of hypomethylation associated with clinical measures of photo-aging. We replicate these findings using whole genome bisulfite sequencing, comparing epidermis from an additional set of younger and older subjects. These blocks largely overlap known hypomethylated blocks in colon cancer and we observe that these same regions are similarly hypomethylated in squamous cell carcinoma samples.
Conclusions: These data implicate large scale epigenomic change in mediating the effects of environmental damage with photo-aging.
Figures


References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

