Assessing structural variation in a personal genome-towards a human reference diploid genome
- PMID: 25886820
- PMCID: PMC4490614
- DOI: 10.1186/s12864-015-1479-3
Assessing structural variation in a personal genome-towards a human reference diploid genome
Abstract
Background: Characterizing large genomic variants is essential to expanding the research and clinical applications of genome sequencing. While multiple data types and methods are available to detect these structural variants (SVs), they remain less characterized than smaller variants because of SV diversity, complexity, and size. These challenges are exacerbated by the experimental and computational demands of SV analysis. Here, we characterize the SV content of a personal genome with Parliament, a publicly available consensus SV-calling infrastructure that merges multiple data types and SV detection methods.
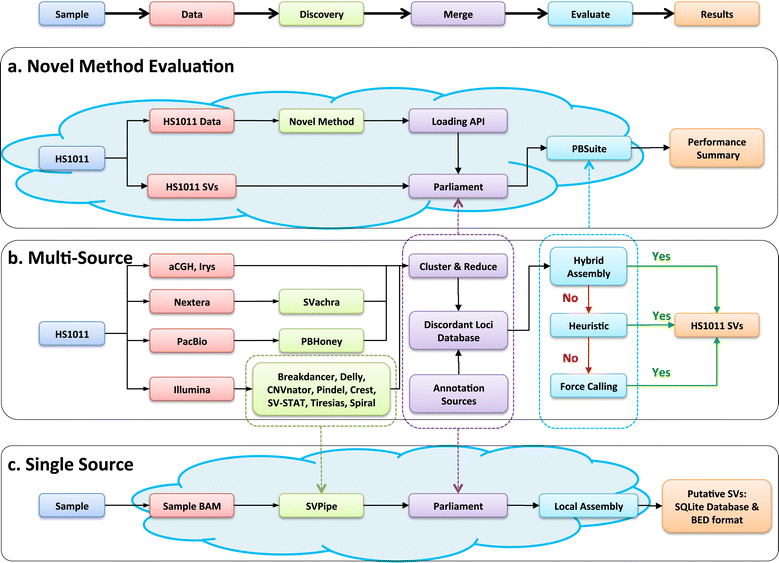
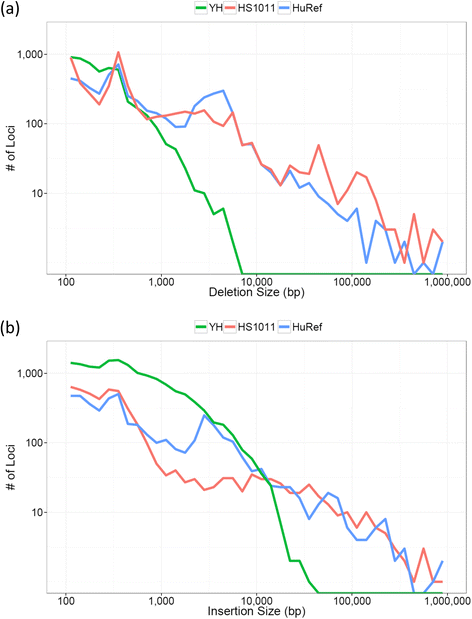
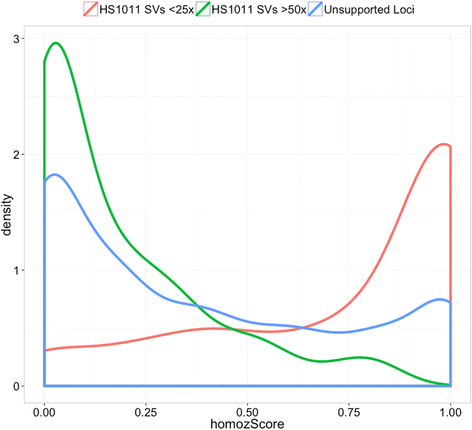
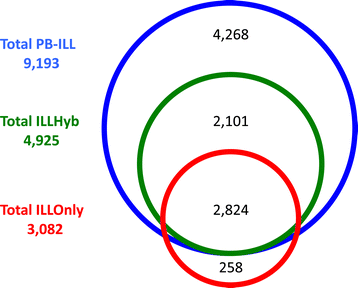
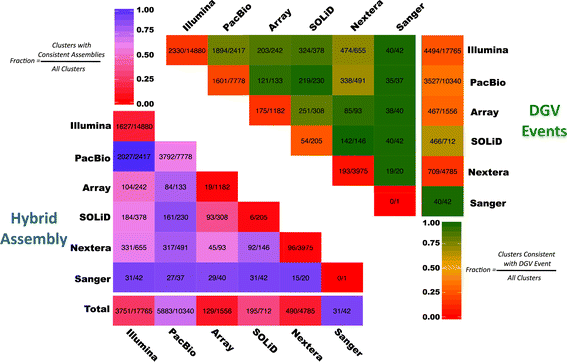
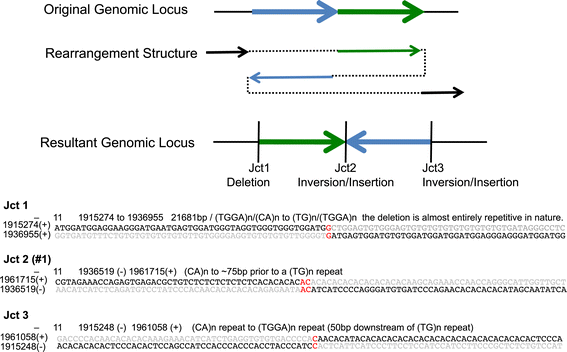
Results: We demonstrate Parliament's efficacy via integrated analyses of data from whole-genome array comparative genomic hybridization, short-read next-generation sequencing, long-read (Pacific BioSciences RSII), long-insert (Illumina Nextera), and whole-genome architecture (BioNano Irys) data from the personal genome of a single subject (HS1011). From this genome, Parliament identified 31,007 genomic loci between 100 bp and 1 Mbp that are inconsistent with the hg19 reference assembly. Of these loci, 9,777 are supported as putative SVs by hybrid local assembly, long-read PacBio data, or multi-source heuristics. These SVs span 59 Mbp of the reference genome (1.8%) and include 3,801 events identified only with long-read data. The HS1011 data and complete Parliament infrastructure, including a BAM-to-SV workflow, are available on the cloud-based service DNAnexus.
Conclusions: HS1011 SV analysis reveals the limits and advantages of multiple sequencing technologies, specifically the impact of long-read SV discovery. With the full Parliament infrastructure, the HS1011 data constitute a public resource for novel SV discovery, software calibration, and personal genome structural variation analysis.
Figures

References
-
- Boerwinkle E, Heckbert SR. Following-Up Genome-Wide Association Study Signals Lessons Learned From Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Targeted Sequencing Study. Circ Cardiovasc Genet. 2014;7:332–4. doi: 10.1161/CIRCGENETICS.113.000078. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

