Sampling with poling-based flux balance analysis: optimal versus sub-optimal flux space analysis of Actinobacillus succinogenes
- PMID: 25887116
- PMCID: PMC4350952
- DOI: 10.1186/s12859-015-0476-5
Sampling with poling-based flux balance analysis: optimal versus sub-optimal flux space analysis of Actinobacillus succinogenes
Abstract
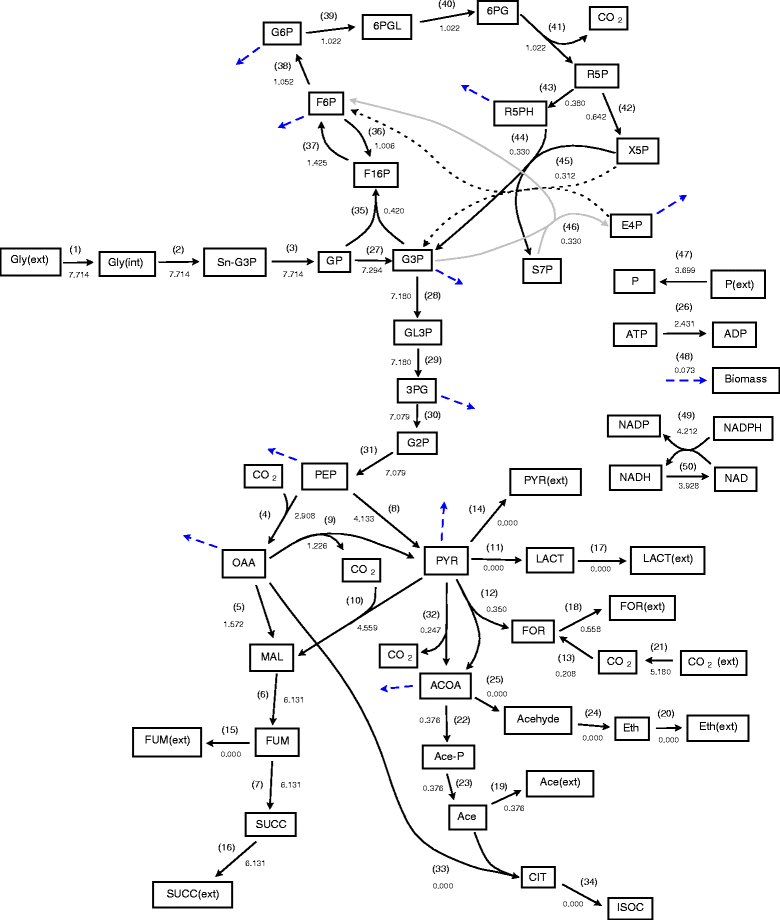
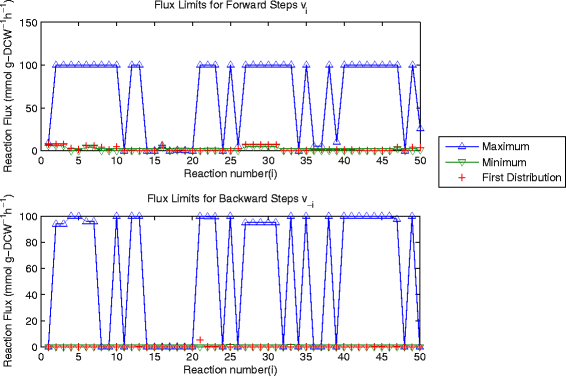
Background: Flux balance analysis is traditionally implemented to identify the maximum theoretical flux for some specified reaction and a single distribution of flux values for all the reactions present which achieve this maximum value. However it is well known that the uncertainty in reaction networks due to branches, cycles and experimental errors results in a large number of combinations of internal reaction fluxes which can achieve the same optimal flux value.
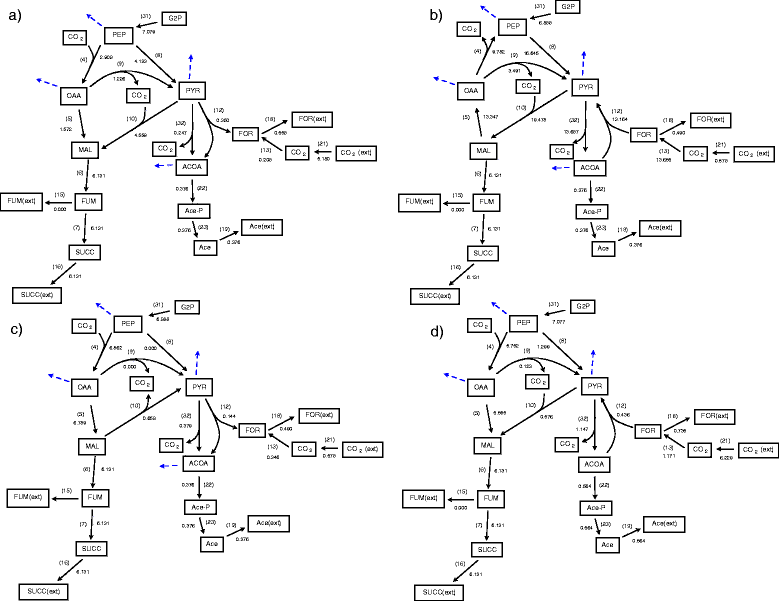
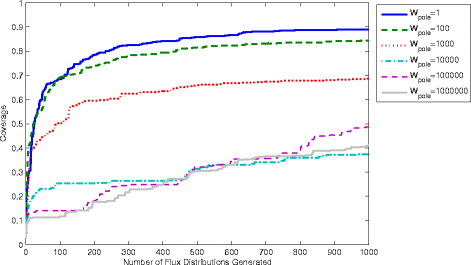
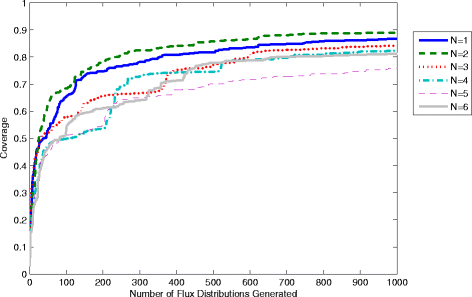
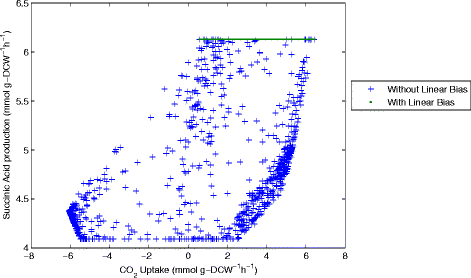
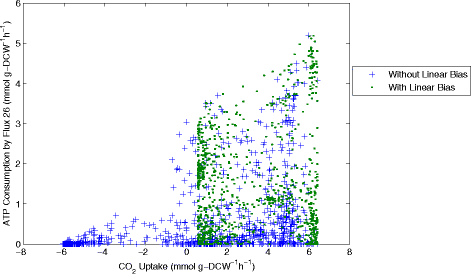
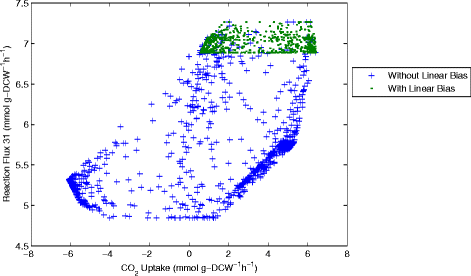
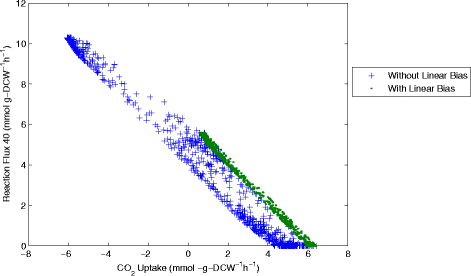
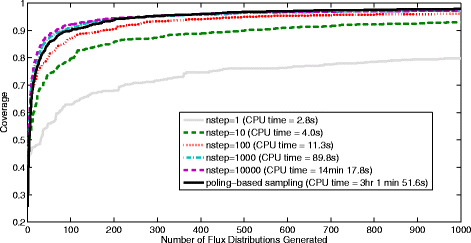
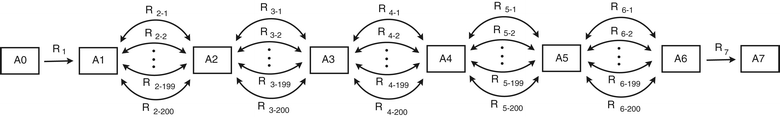
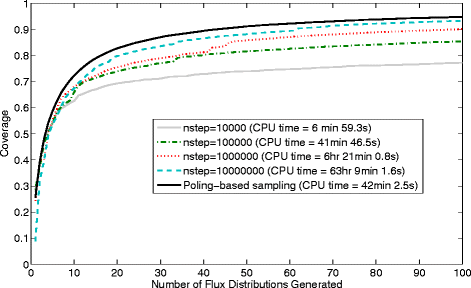
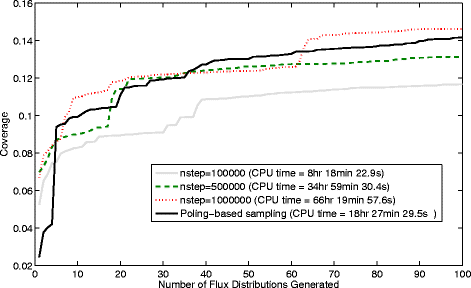
Results: In this work, we have modified the applied linear objective of flux balance analysis to include a poling penalty function, which pushes each new set of reaction fluxes away from previous solutions generated. Repeated poling-based flux balance analysis generates a sample of different solutions (a characteristic set), which represents all the possible functionality of the reaction network. Compared to existing sampling methods, for the purpose of generating a relatively "small" characteristic set, our new method is shown to obtain a higher coverage than competing methods under most conditions. The influence of the linear objective function on the sampling (the linear bias) constrains optimisation results to a subspace of optimal solutions all producing the same maximal fluxes. Visualisation of reaction fluxes plotted against each other in 2 dimensions with and without the linear bias indicates the existence of correlations between fluxes. This method of sampling is applied to the organism Actinobacillus succinogenes for the production of succinic acid from glycerol.
Conclusions: A new method of sampling for the generation of different flux distributions (sets of individual fluxes satisfying constraints on the steady-state mass balances of intermediates) has been developed using a relatively simple modification of flux balance analysis to include a poling penalty function inside the resulting optimisation objective function. This new methodology can achieve a high coverage of the possible flux space and can be used with and without linear bias to show optimal versus sub-optimal solution spaces. Basic analysis of the Actinobacillus succinogenes system using sampling shows that in order to achieve the maximal succinic acid production CO₂ must be taken into the system. Solutions involving release of CO₂ all give sub-optimal succinic acid production.
Figures





References
-
- Binns M, Theodoropoulos C. An integrated knowledge-based approach for modeling biochemical reaction networks. Comput Chem Eng. 2011;35:3025–43. doi: 10.1016/j.compchemeng.2011.03.030. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

