Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations
- PMID: 25887147
- PMCID: PMC4381444
- DOI: 10.1186/s13059-015-0612-6
Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations
Abstract
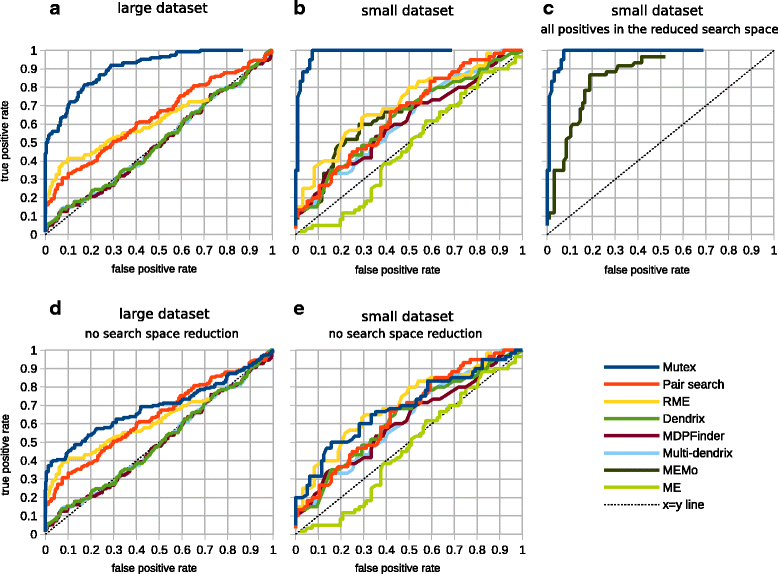
We present a novel method for the identification of sets of mutually exclusive gene alterations in a given set of genomic profiles. We scan the groups of genes with a common downstream effect on the signaling network, using a mutual exclusivity criterion that ensures that each gene in the group significantly contributes to the mutual exclusivity pattern. We test the method on all available TCGA cancer genomics datasets, and detect multiple previously unreported alterations that show significant mutual exclusivity and are likely to be driver events.
Figures
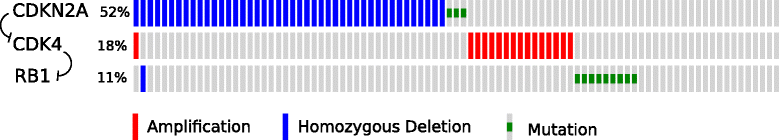
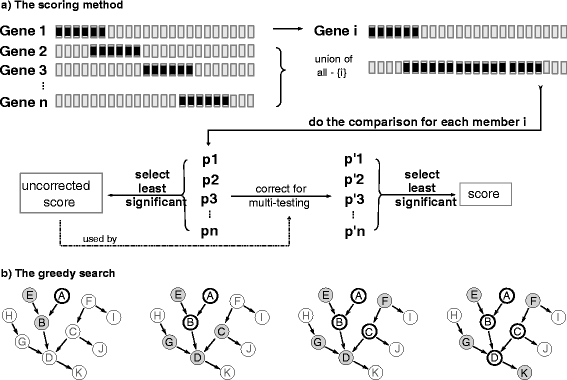
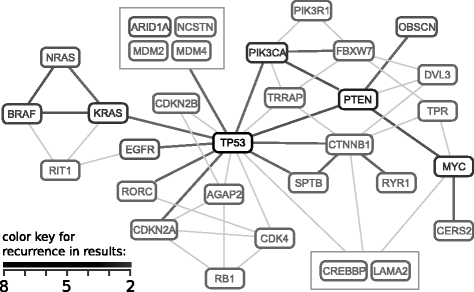
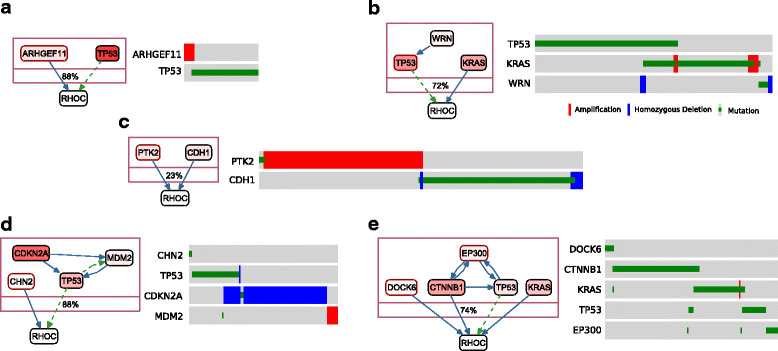
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

