The ensembl regulatory build
- PMID: 25887522
- PMCID: PMC4407537
- DOI: 10.1186/s13059-015-0621-5
The ensembl regulatory build
Abstract
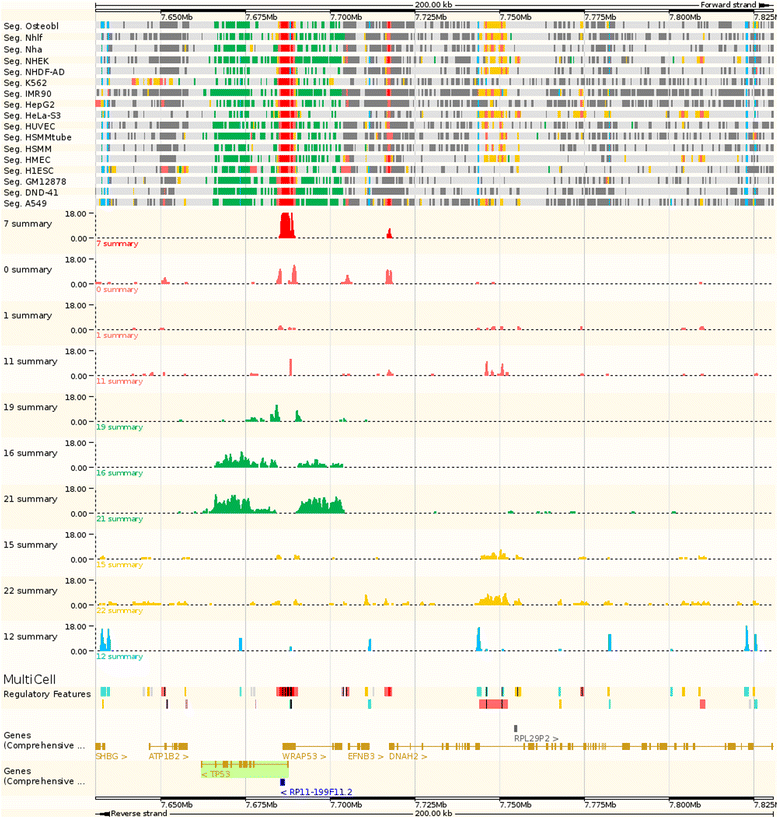
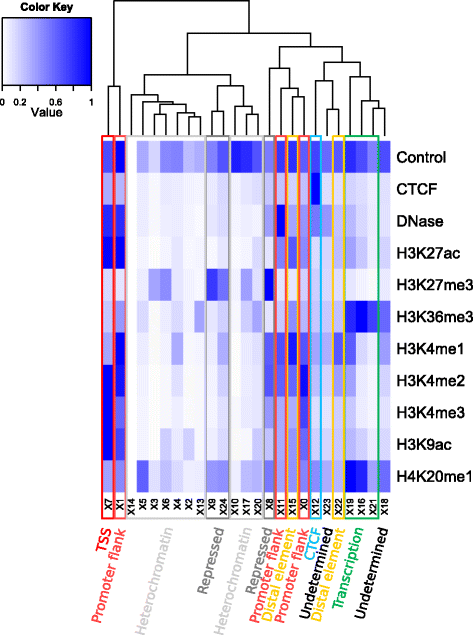
Most genomic variants associated with phenotypic traits or disease do not fall within gene coding regions, but in regulatory regions, rendering their interpretation difficult. We collected public data on epigenetic marks and transcription factor binding in human cell types and used it to construct an intuitive summary of regulatory regions in the human genome. We verified it against independent assays for sensitivity. The Ensembl Regulatory Build will be progressively enriched when more data is made available. It is freely available on the Ensembl browser, from the Ensembl Regulation MySQL database server and in a dedicated track hub.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

