Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis
- PMID: 25887579
- PMCID: PMC4376514
- DOI: 10.1186/s12864-015-1433-4
Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis
Abstract
Background: The fruit of litchi (Litchi chinensis) comprises a white translucent edible aril surrounded by a pericarp. The pericarp of litchi has been the focus of studies associated with fruit size, coloration, cracking and shelf life. However, research at the molecular level has been limited by the lack of genomic and transcriptomic information. In this study, an analysis of the transcriptome of litchi pericarp was performed to obtain information regarding the molecular mechanisms underlying the physiological changes in the pericarp, including those leading to fruit surface coloration.
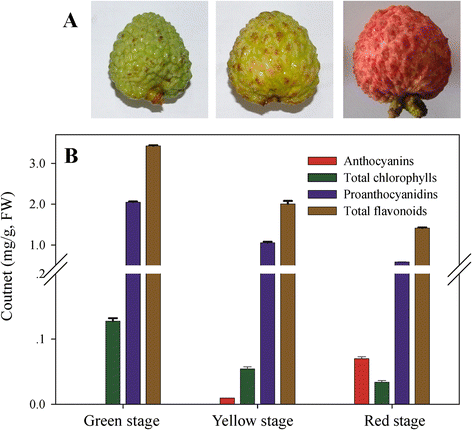
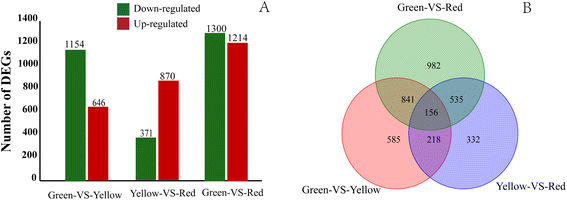
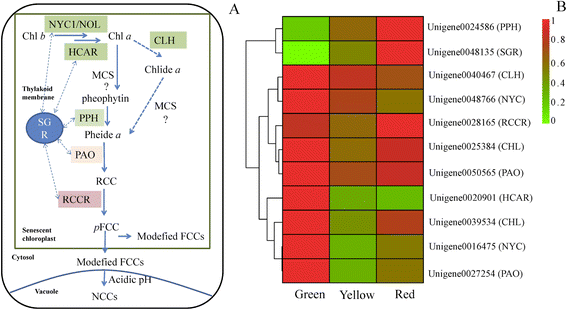
Results: Coincident with the rapid break down of chlorophyll, but substantial increase of anthocyanins in litchi pericarp as fruit developed, two major physiological changes, degreening and pigmentation were visually apparent. In this study, a cDNA library of litchi pericarp with three different coloration stages was constructed. A total of 4.7 Gb of raw RNA-Seq data was generated and this was then de novo assembled into 51,089 unigenes with a mean length of 737 bp. Approximately 70% of the unigenes (34,705) could be annotated based on public protein databases and, of these, 3,649 genes were significantly differentially expressed between any two coloration stages, while 156 genes were differentially expressed among all three stages. Genes encoding enzymes involved in chlorophyll degradation and flavonoid biosynthesis were identified in the transcriptome dataset. The transcript expression patterns of the Stay Green (SGR) protein suggested a key role in chlorophyll degradation in the litchi pericarp, and this conclusion was supported by the result of an assay over-expressing LcSGR protein in tobacco leaves. We also found that the expression levels of most genes especially late anthocyanin biosynthesis genes were co-ordinated up-regulated coincident with the accumulation of anthocyanins, and that candidate MYB transcription factors that likely regulate flavonoid biosynthesis were identified.
Conclusions: This study provides a large collection of transcripts and expression profiles associated with litchi fruit maturation processes, including coloration. Since most of the unigenes were annotated, they provide a platform for litchi functional genomic research within this species.
Figures







Similar articles
-
Three LcABFs are Involved in the Regulation of Chlorophyll Degradation and Anthocyanin Biosynthesis During Fruit Ripening in Litchi chinensis.Plant Cell Physiol. 2019 Feb 1;60(2):448-461. doi: 10.1093/pcp/pcy219. Plant Cell Physiol. 2019. PMID: 30407601
-
Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of Litchi chinensis Sonn.PLoS One. 2011 Apr 29;6(4):e19455. doi: 10.1371/journal.pone.0019455. PLoS One. 2011. PMID: 21559331 Free PMC article.
-
Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi.Plant Physiol. 2023 Jul 3;192(3):1913-1927. doi: 10.1093/plphys/kiad118. Plant Physiol. 2023. PMID: 36843134 Free PMC article.
-
Sugar Transport, Metabolism and Signaling in Fruit Development of Litchi chinensis Sonn: A Review.Int J Mol Sci. 2021 Oct 18;22(20):11231. doi: 10.3390/ijms222011231. Int J Mol Sci. 2021. PMID: 34681891 Free PMC article. Review.
-
Combinatorial approaches for controlling pericarp browning in Litchi (Litchi chinensis) fruit.J Food Sci Technol. 2015 Sep;52(9):5418-26. doi: 10.1007/s13197-015-1712-8. Epub 2015 Feb 11. J Food Sci Technol. 2015. PMID: 26344958 Free PMC article. Review.
Cited by
-
Residue Analysis and the Effect of Preharvest Forchlorfenuron (CPPU) Application on On-Tree Quality Maintenance of Ripe Fruit in "Feizixiao" Litchi (Litchi chinensis Sonn.).Front Plant Sci. 2022 Mar 4;13:829635. doi: 10.3389/fpls.2022.829635. eCollection 2022. Front Plant Sci. 2022. PMID: 35310679 Free PMC article.
-
Development of molecular markers based on the promoter difference of LcFT1 to discriminate easy- and difficult-flowering litchi germplasm resources and its application in crossbreeding.BMC Plant Biol. 2021 Nov 16;21(1):539. doi: 10.1186/s12870-021-03309-7. BMC Plant Biol. 2021. PMID: 34784881 Free PMC article.
-
Natural Variation Underlies Differences in ETHYLENE RESPONSE FACTOR17 Activity in Fruit Peel Degreening.Plant Physiol. 2018 Mar;176(3):2292-2304. doi: 10.1104/pp.17.01320. Epub 2018 Feb 5. Plant Physiol. 2018. PMID: 29431631 Free PMC article.
-
Transcriptome changes between compatible and incompatible graft combination of Litchi chinensis by digital gene expression profile.Sci Rep. 2017 Jun 21;7(1):3954. doi: 10.1038/s41598-017-04328-x. Sci Rep. 2017. PMID: 28638079 Free PMC article.
-
The MAPKKK and MAPKK gene families in banana: identification, phylogeny and expression during development, ripening and abiotic stress.Sci Rep. 2017 Apr 25;7(1):1159. doi: 10.1038/s41598-017-01357-4. Sci Rep. 2017. PMID: 28442729 Free PMC article.
References
-
- Andersen Ø, Jordheim M. The anthocyanins. In: Andersen ØM, Markham KR, editors. Flavonoids. Boca Raton: CRC Press; 2006. pp. 471–551.
-
- Wang HC, Huang XM, Hu GB, Yang Z, Huang HB. A comparative study of chlorophyll loss and its related mechanism during fruit maturation in the pericarp of fast- and slow-degreening litchi pericarp. Sci Hortic. 2005;106(2):247–57. doi: 10.1016/j.scienta.2005.03.007. - DOI
-
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andres CB, et al. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell. 2012;24(2):507–18. doi: 10.1105/tpc.111.089474. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

