Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation
- PMID: 25887697
- PMCID: PMC4369366
- DOI: 10.1186/s12864-015-1378-7
Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation
Abstract
Background: The animal gastrointestinal tract contains a complex community of microbes, whose composition ultimately reflects the co-evolution of microorganisms with their animal host and the diet adopted by the host. Although the importance of gut microbiota of humans has been well demonstrated, there is a paucity of research regarding non-human primates (NHPs), especially herbivorous NHPs.
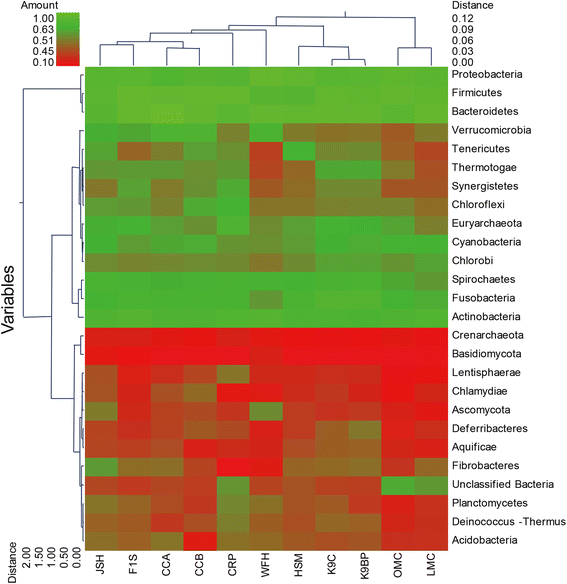
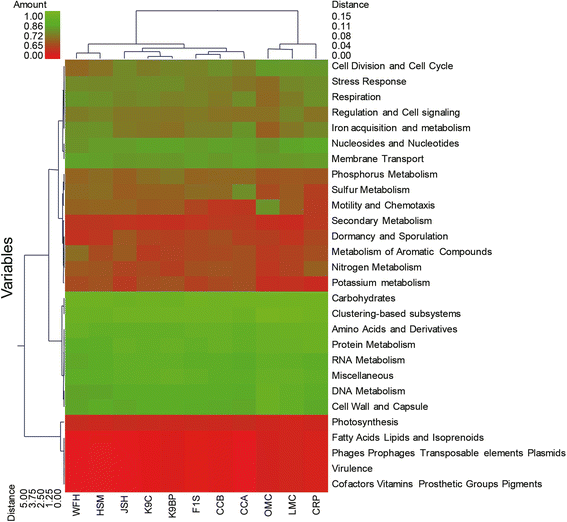
Results: In this study, an analysis of 97,942 pyrosequencing reads generated from Rhinopithecus bieti fecal DNA extracts was performed to help better understanding of the microbial diversity and functional capacity of the R. bieti gut microbiome. The taxonomic analysis of the metagenomic reads indicated that R. bieti fecal microbiomes were dominated by Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria phyla. The comparative analysis of taxonomic classification revealed that the metagenome of R. bieti was characterized by an overrepresentation of bacteria of phylum Fibrobacteres and Spirochaetes as compared with other animals. Primary functional categories were associated mainly with protein, carbohydrates, amino acids, DNA and RNA metabolism, cofactors, cell wall and capsule and membrane transport. Comparing glycoside hydrolase profiles of R. bieti with those of other animal revealed that the R. bieti microbiome was most closely related to cow rumen.
Conclusions: Metagenomic and functional analysis demonstrated that R. bieti possesses a broad diversity of bacteria and numerous glycoside hydrolases responsible for lignocellulosic biomass degradation which might reflect the adaptations associated with a diet rich in fibrous matter. These results would contribute to the limited body of NHPs metagenome studies and provide a unique genetic resource of plant cell wall degrading microbial enzymes. However, future studies on the metagenome sequencing of R. bieti regarding the effects of age, genetics, diet and environment on the composition and activity of the metagenomes are required.
Figures


References
-
- Long YC, Kirkpatrick CR, Zhong T, Xiao L. Report on distribution, population and ecology of the Yunnan snub-nosed monkey (Rhinopithecus bieti) Primates. 1994;35:241–250. doi: 10.1007/BF02382060. - DOI
-
- Bennett EL, Davies AG. The ecology of Asian colobines. In: Davies AG, Oates JF, editors. Colobine monkeys: Their ecology, behaviour and evolution. Cambridge, UK: Cambridge University Press; 1994. pp. 129–171.
-
- Peng YZ, Zhang YP, Ye ZZ, Liu RL. Study on the stomachs in three species of snub-nosed monkeys. Zoonoses Res. 1983;4:167–175.
-
- Chen JJ, Lu T, Liu JS, Huang ZR. Observations on the stomach of Rhinopithecus Roxellanae. Acta Theriologica Sinica. 1995;15:176–180.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

