Measurably evolving pathogens in the genomic era
- PMID: 25887947
- PMCID: PMC4457702
- DOI: 10.1016/j.tree.2015.03.009
Measurably evolving pathogens in the genomic era
Abstract
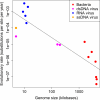
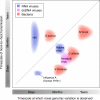
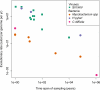
Current sequencing technologies have created unprecedented opportunities for studying microbial populations. For pathogens with comparatively low per-site mutation rates, such as DNA viruses and bacteria, whole-genome sequencing can reveal the accumulation of novel genetic variation between population samples taken at different times. The concept of 'measurably evolving populations' and related analytical approaches have provided powerful insights for fast-evolving RNA viruses, but their application to other pathogens is still in its infancy. We argue that previous distinctions between slow- and fast-evolving pathogens become blurred once evolution is assessed at a genome-wide scale, and we highlight important analytical challenges to be overcome to infer pathogen population dynamics from genomic data.
Keywords: DNA virus; bacteria; epidemiological models; evolutionary rate; infectious disease; phylodynamics.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
References
-
- Drummond AJ, et al. Measurably evolving populations. Trends Ecol. Evol. 2003;18:481–488.
-
- Grenfell BT, et al. Unifying the Epidemiological and Evolutionary Dynamics of Pathogens. Science. 2004;303:327–332. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

