Evolution of lysine acetylation in the RNA polymerase II C-terminal domain
- PMID: 25887984
- PMCID: PMC4362643
- DOI: 10.1186/s12862-015-0327-z
Evolution of lysine acetylation in the RNA polymerase II C-terminal domain
Abstract
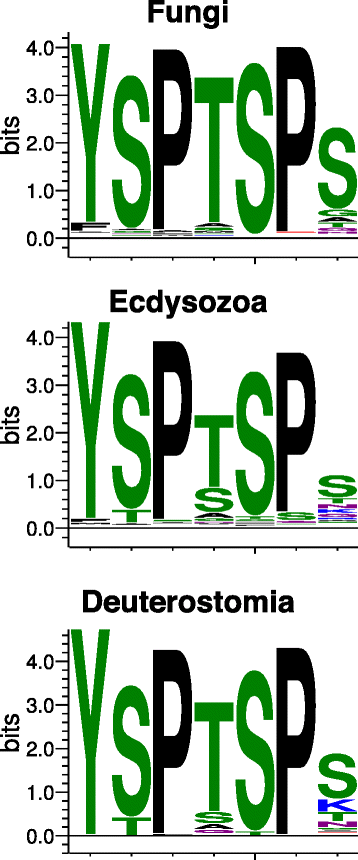
Background: RPB1, the largest subunit of RNA polymerase II, contains a highly modifiable C-terminal domain (CTD) that consists of variations of a consensus heptad repeat sequence (Y1S2P3T4S5P6S7). The consensus CTD repeat motif and tandem organization represent the ancestral state of eukaryotic RPB1, but across eukaryotes CTDs show considerable diversity in repeat organization and sequence content. These differences may reflect lineage-specific CTD functions mediated by protein interactions. Mammalian CTDs contain eight non-consensus repeats with a lysine in the seventh position (K7). Posttranslational acetylation of these sites was recently shown to be required for proper polymerase pausing and regulation of two growth factor-regulated genes.
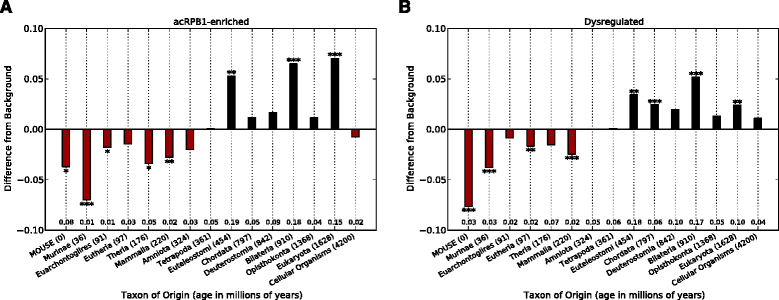
Results: To investigate the origins and function of RPB1 CTD acetylation (acRPB1), we computationally reconstructed the evolution of the CTD repeat sequence across eukaryotes and analyzed the evolution and function of genes dysregulated when acRPB1 is disrupted. Modeling the evolutionary dynamics of CTD repeat count and sequence content across diverse eukaryotes revealed an expansion of the CTD in the ancestors of Metazoa. The new CTD repeats introduced the potential for acRPB1 due to the appearance of distal repeats with lysine at position seven. This was followed by a further increase in the number of lysine-containing repeats in developmentally complex clades like Deuterostomia. Mouse genes enriched for acRPB1 occupancy at their promoters and genes with significant expression changes when acRPB1 is disrupted are enriched for several functions, such as growth factor response, gene regulation, cellular adhesion, and vascular development. Genes occupied and regulated by acRPB1 show significant enrichment for evolutionary origins in the early history of eukaryotes through early vertebrates.
Conclusions: Our combined functional and evolutionary analyses show that RPB1 CTD acetylation was possible in the early history of animals, and that the K7 content of the CTD expanded in specific developmentally complex metazoan lineages. The functional analysis of genes regulated by acRPB1 highlight functions involved in the origin of and diversification of complex Metazoa. This suggests that acRPB1 may have played a role in the success of animals.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

