A haploproficient interaction of the transaldolase paralogue NQM1 with the transcription factor VHR1 affects stationary phase survival and oxidative stress resistance
- PMID: 25887987
- PMCID: PMC4331311
- DOI: 10.1186/s12863-015-0171-6
A haploproficient interaction of the transaldolase paralogue NQM1 with the transcription factor VHR1 affects stationary phase survival and oxidative stress resistance
Abstract
Background: Studying the survival of yeast in stationary phase, known as chronological lifespan, led to the identification of molecular ageing factors conserved from yeast to higher organisms. To identify functional interactions among yeast chronological ageing genes, we conducted a haploproficiency screen on the basis of previously identified long-living mutants. For this, we created a library of heterozygous Saccharomyces cerevisiae double deletion strains and aged them in a competitive manner.
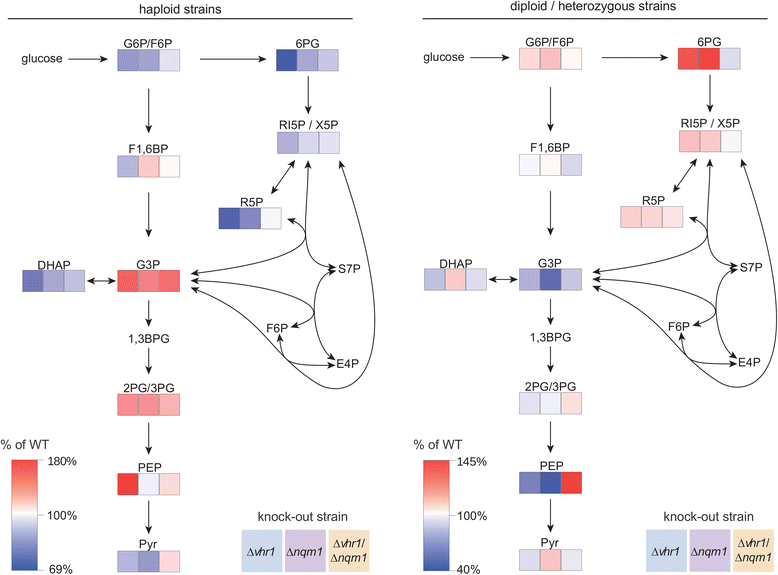
Results: Stationary phase survival was prolonged in a double heterozygous mutant of the metabolic enzyme non-quiescent mutant 1 (NQM1), a paralogue to the pentose phosphate pathway enzyme transaldolase (TAL1), and the transcription factor vitamin H response transcription factor 1 (VHR1). We find that cells deleted for the two genes possess increased clonogenicity at late stages of stationary phase survival, but find no indication that the mutations delay initial mortality upon reaching stationary phase, canonically defined as an extension of chronological lifespan. We show that both genes influence the concentration of metabolites of glycolysis and the pentose phosphate pathway, central metabolic players in the ageing process, and affect osmolality of growth media in stationary phase cultures. Moreover, NQM1 is glucose repressed and induced in a VHR1 dependent manner upon caloric restriction, on non-fermentable carbon sources, as well as under osmotic and oxidative stress. Finally, deletion of NQM1 is shown to confer resistance to oxidizing substances.
Conclusions: The transaldolase paralogue NQM1 and the transcription factor VHR1 interact haploproficiently and affect yeast stationary phase survival. The glucose repressed NQM1 gene is induced under various stress conditions, affects stress resistance and this process is dependent on VHR1. While NQM1 appears not to function in the pentose phosphate pathway, the interplay of NQM1 with VHR1 influences the yeast metabolic homeostasis and stress tolerance during stationary phase, processes associated with yeast ageing.
Figures





Similar articles
-
Characterization of non-oxidative transaldolase and transketolase enzymes in the pentose phosphate pathway with regard to xylose utilization by recombinant Saccharomyces cerevisiae.Enzyme Microb Technol. 2012 Jun 10;51(1):16-25. doi: 10.1016/j.enzmictec.2012.03.008. Epub 2012 Apr 4. Enzyme Microb Technol. 2012. PMID: 22579386
-
Molecular analysis of the structural gene for yeast transaldolase.Eur J Biochem. 1990 Mar 30;188(3):597-603. doi: 10.1111/j.1432-1033.1990.tb15440.x. Eur J Biochem. 1990. PMID: 2185015
-
Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress.Mol Gen Genet. 1996 Sep 25;252(4):456-64. doi: 10.1007/BF02173011. Mol Gen Genet. 1996. PMID: 8879247
-
The Pentose Phosphate Pathway in Yeasts-More Than a Poor Cousin of Glycolysis.Biomolecules. 2021 May 12;11(5):725. doi: 10.3390/biom11050725. Biomolecules. 2021. PMID: 34065948 Free PMC article. Review.
-
Oxidative stress, inflammation and carcinogenesis are controlled through the pentose phosphate pathway by transaldolase.Trends Mol Med. 2011 Jul;17(7):395-403. doi: 10.1016/j.molmed.2011.01.014. Epub 2011 Mar 2. Trends Mol Med. 2011. PMID: 21376665 Free PMC article. Review.
Cited by
-
Genome-Wide Mutant Screening in Yeast Reveals that the Cell Wall is a First Shield to Discriminate Light From Heavy Lanthanides.Front Microbiol. 2022 May 19;13:881535. doi: 10.3389/fmicb.2022.881535. eCollection 2022. Front Microbiol. 2022. PMID: 35663896 Free PMC article.
-
Restoring fertility in yeast hybrids: Breeding and quantitative genetics of beneficial traits.Proc Natl Acad Sci U S A. 2021 Sep 21;118(38):e2101242118. doi: 10.1073/pnas.2101242118. Proc Natl Acad Sci U S A. 2021. PMID: 34518218 Free PMC article.
References
-
- Krüger A, Vowinckel J, Mülleder M, Grote P, Capuano F, Bluemlein K, et al. Tpo1-mediated spermine and spermidine export controls cell cycle delay and times antioxidant protein expression during the oxidative stress response. EMBO Rep. 2013;14:1113–9. doi: 10.1038/embor.2013.165. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

