Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions
- PMID: 25887988
- PMCID: PMC4399226
- DOI: 10.1186/s12711-014-0074-4
Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions
Abstract
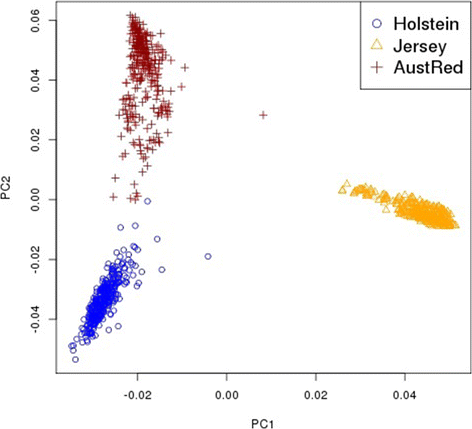
Background: Genomic selection is increasingly widely practised, particularly in dairy cattle. However, the accuracy of current predictions using GBLUP (genomic best linear unbiased prediction) decays rapidly across generations, and also as selection candidates become less related to the reference population. This is likely caused by the effects of causative mutations being dispersed across many SNPs (single nucleotide polymorphisms) that span large genomic intervals. In this paper, we hypothesise that the use of a nonlinear method (BayesR), combined with a multi-breed (Holstein/Jersey) reference population will map causative mutations with more precision than GBLUP and this, in turn, will increase the accuracy of genomic predictions for selection candidates that are less related to the reference animals.
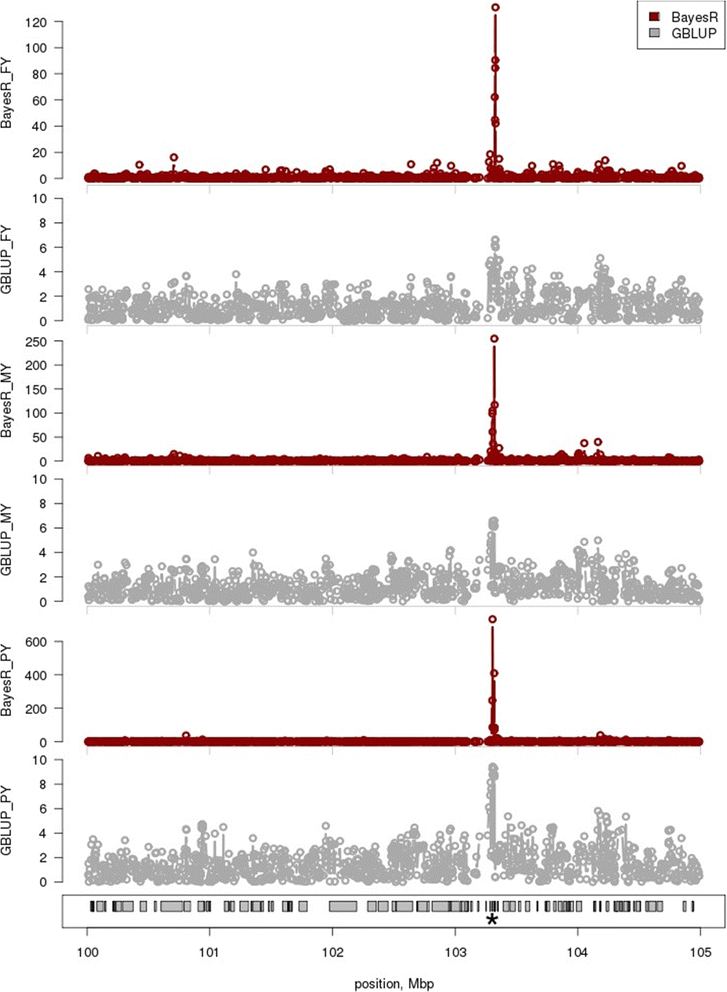
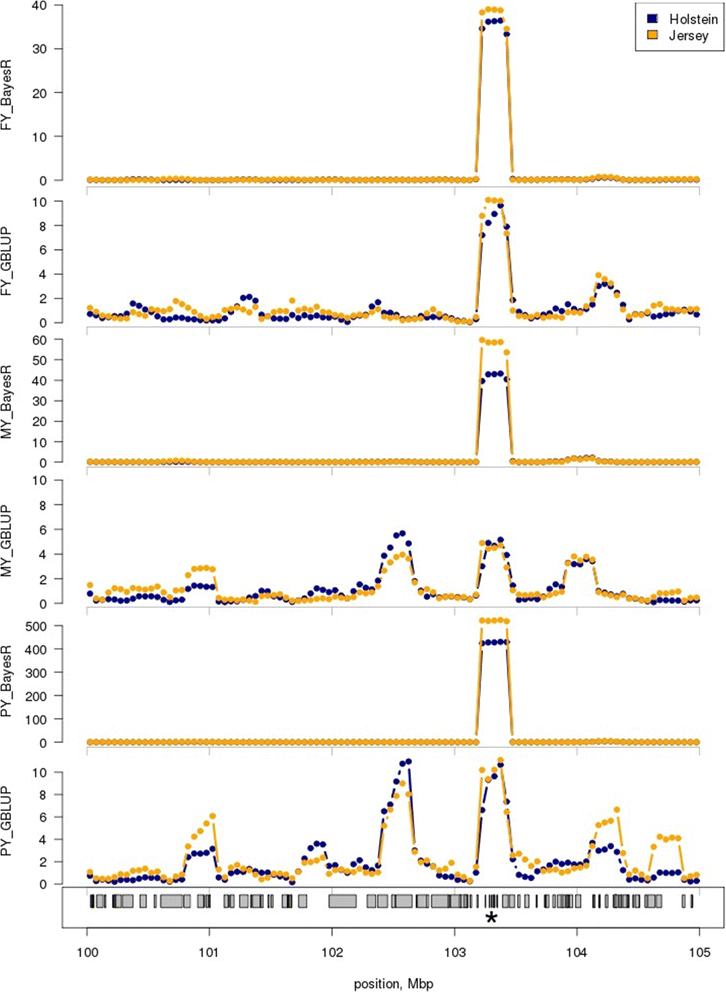
Results: BayesR improved the across-breed prediction accuracy for Australian Red dairy cattle for five milk yield and composition traits by an average of 7% over the GBLUP approach (Australian Red animals were not included in the reference population). Using the multi-breed reference population with BayesR improved accuracy of prediction in Australian Red cattle by 2 - 5% compared to using BayesR with a single breed reference population. Inclusion of 8478 Holstein and 3917 Jersey cows in the reference population improved accuracy of predictions for these breeds by 4 and 5%. However, predictions for Holstein and Jersey cattle were similar using within-breed and multi-breed reference populations. We propose that the improvement in across-breed prediction achieved by BayesR with the multi-breed reference population is due to more precise mapping of quantitative trait loci (QTL), which was demonstrated for several regions. New candidate genes with functional links to milk synthesis were identified using differential gene expression in the mammary gland.
Conclusions: QTL detection and genomic prediction are usually considered independently but persistence of genomic prediction accuracies across breeds requires accurate estimation of QTL effects. We show that accuracy of across-breed genomic predictions was higher with BayesR than with GBLUP and that BayesR mapped QTL more precisely. Further improvements of across-breed accuracy of genomic predictions and QTL mapping could be achieved by increasing the size of the reference population, including more breeds, and possibly by exploiting pleiotropic effects to improve mapping efficiency for QTL with small effects.
Figures
References
-
- Kachman SD, Spangler ML, Bennett GL, Hanford KJ, Kuehn LA, Snelling WM, Thallman RM, Saatchi M, Garrick DJ, Schnabel RD, Taylor JF, Pollak EJ. Comparison of molecular breeding values based on within- and across-breed training in beef cattle. Genet Sel Evol. 2013;45:30. doi: 10.1186/1297-9686-45-30. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

