An efficient method for zoospore production, infection and real-time quantification of Phytophthora cajani causing Phytophthora blight disease in pigeonpea under elevated atmospheric CO₂
- PMID: 25888001
- PMCID: PMC4377013
- DOI: 10.1186/s12870-015-0470-0
An efficient method for zoospore production, infection and real-time quantification of Phytophthora cajani causing Phytophthora blight disease in pigeonpea under elevated atmospheric CO₂
Abstract
Background: Phytophthora blight caused by Phytophthora cajani is an emerging disease of pigeonpea (Cajanus cajan L.) affecting the crop irrespective of cropping system, cultivar grown and soil types. Current detection and identification methods for Phytophthora species rely primarily on cultural and morphological characteristics, the assessment of which is time-consuming and not always suitable. Sensitive and reliable methods for isolation, identification, zoospore production and estimating infection severity are therefore desirable in case of Phytophthora blight of pigeonpea.
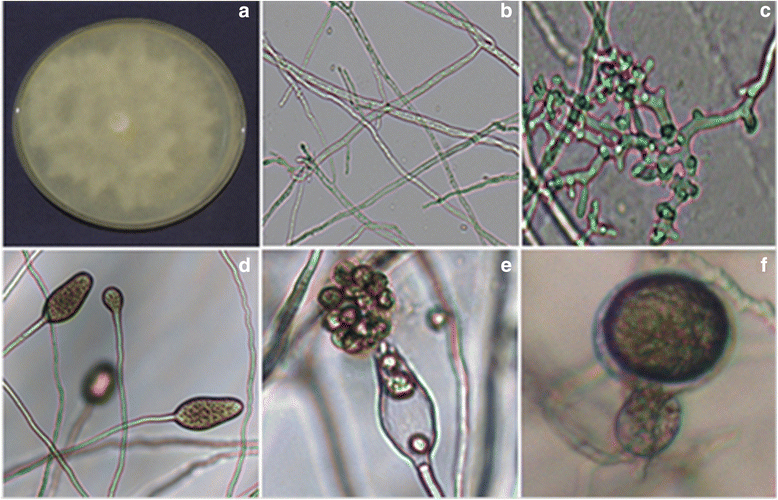
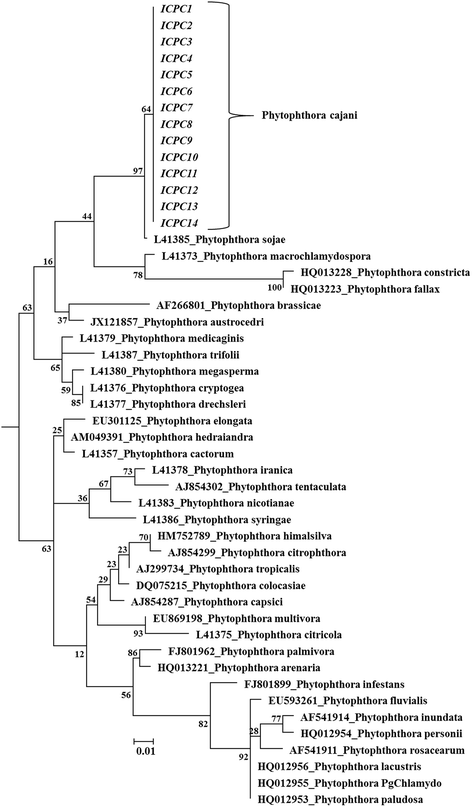
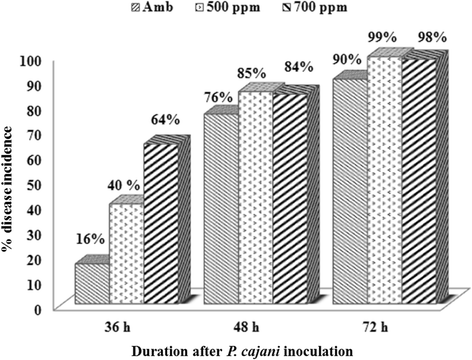
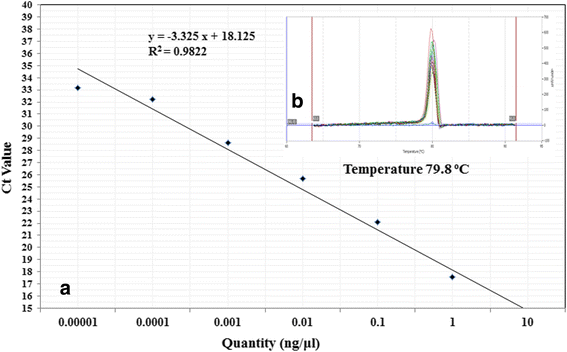
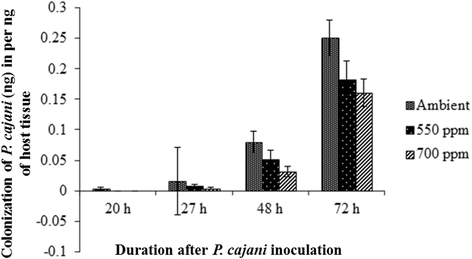
Results: In this study, protocols for isolation and identification of Phytophthora blight of pigeonpea were standardized. Also the method for zoospore production and in planta infection of P. cajani was developed. Quantification of fungal colonization by P. cajani using real-time PCR was further standardized. Phytophthora species infecting pigeonpea was identified based on mycological characters such as growth pattern, mycelium structure and sporangial morphology of the isolates and confirmed through molecular characterization (sequence deposited in GenBank). For Phytophthora disease development, zoospore suspension of 1 × 10(5) zoospores per ml was found optimum. Phytophthora specific real-time PCR assay was developed using specific primers based on internal transcribed spacer (ITS) 1 and 2. Use of real-time PCR allowed the quantitative estimation of fungal biomass in plant tissues. Detection sensitivities were within the range of 0.001 pg fungal DNA. A study to see the effect of elevated CO₂ on Phytophthora blight incidence was also conducted which indicated no significant difference in disease incidence, but incubation period delayed under elevated CO₂ as compared to ambient level.
Conclusion: The zoospore infection method for Phytophthora blight of pigeonpea will facilitate the small and large scale inoculation experiments and thus devise a platform for rapid and reliable screening against Phytophthora blight disease of pigeonpea. qPCR allowed a reliable detection and quantification of P. cajani in samples with low pathogen densities. This can be useful in early warning systems prior to potential devastating outbreak of the disease.
Figures
References
-
- FAO. FAO Year book (Production). Food and Agriculture Organization of the United Nations. 2012.
-
- Pande S, Sharma M, Mangla UN, Ghosh R, Sundaresan G. Phytophthora blight of Pigeonpea [Cajanus cajan (L.) Millsp.]: an updating review of biology, pathogenicity and disease management. Crop Prot. 2011;30:951–7. doi: 10.1016/j.cropro.2011.03.031. - DOI
-
- Sharma M, Pande S, Rao JN, Kumar PA, Reddy DM, Benagi VI, et al. Prevalence of phytophthora blight of pigeonpea in the Deccan Plateau of India. Plant Pathol J. 2006;22:309–13. doi: 10.5423/PPJ.2006.22.4.309. - DOI
-
- Grünwald NJ, Martin FN, Larsen MM, Sullivan CM, Press CM, Coffey MD, et al. Phytophthora-ID.org: a sequence-based Phytophthora identification tool. Plant Dis. 2011;95:337–42. doi: 10.1094/PDIS-08-10-0609. - DOI - PubMed
-
- Cooke DEL, Duncan JM. Phylogenetic analysis of Phytophthora species based on ITS1 and ITS2 sequences of the ribosomal RNA gene repeat. Mycol Res. 1997;6:667–77. doi: 10.1017/S0953756296003218. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

