Annotation of the goat genome using next generation sequencing of microRNA expressed by the lactating mammary gland: comparison of three approaches
- PMID: 25888052
- PMCID: PMC4430871
- DOI: 10.1186/s12864-015-1471-y
Annotation of the goat genome using next generation sequencing of microRNA expressed by the lactating mammary gland: comparison of three approaches
Abstract
Background: MicroRNAs (miRNA) are small endogenous non-coding RNA involved in the post-transcriptional regulation of specific mRNA targets. The first whole goat genome sequence became available in 2013, with few annotations. Our goal was to establish a list of the miRNA expressed in the mammary gland of lactating goats, thus enabling implementation of the goat miRNA repertoire and considerably enriching annotation of the goat genome.
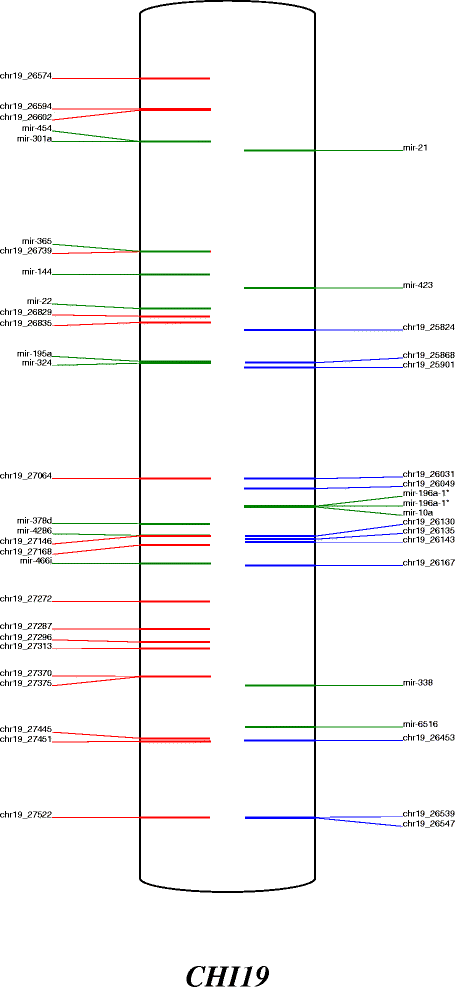
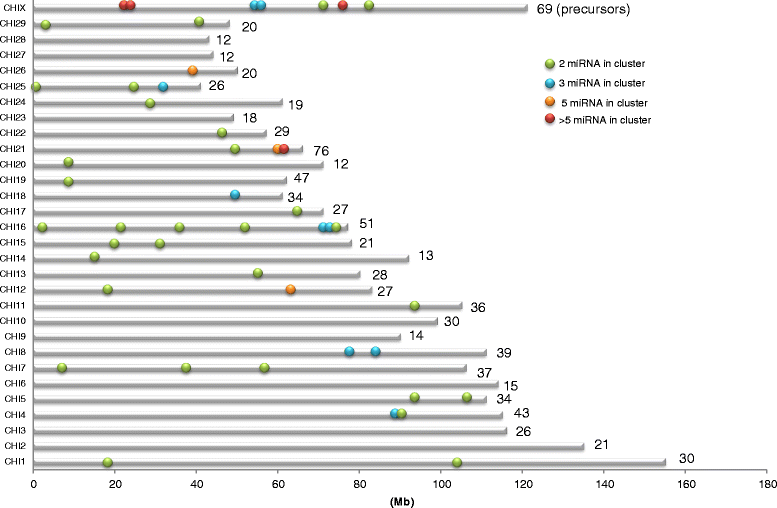
Results: Here, we performed high throughput RNA sequencing on 10 lactating goat mammary glands. The bioinformatic detection of miRNA was carried out using miRDeep2 software. Three different methods were used to predict, quantify and annotate the sequenced reads. The first was a de novo approach based on the prediction of miRNA from the goat genome only. The second approach used bovine miRNA as an external reference whereas the last one used recently available goat miRNA. The three methods enabled the prediction and annotation of hundreds of miRNA, more than 95% were commonly identified. Using bovine miRNA, 1,178 distinct miRNA were detected, together with the annotation of 88 miRNA for which corresponding precursors could not be retrieved in the goat genome, and which were not detected using the de novo approach or with the use of goat miRNA. Each chromosomal coordinate of the precursors determined here were generated and depicted on a reference localisation map. Forty six goat miRNA clusters were also reported. The study revealed 263 precursors located in goat protein-coding genes, amongst which the location of 43 precursors was conserved between human, mouse and bovine, revealing potential new gene regulations in the goat mammary gland. Using the publicly available cattle QTL database, and cow precursors conserved in the goat and expressed in lactating mammary gland, 114 precursors were located within known QTL regions for milk production and composition.
Conclusions: The results reported here represent the first major identification study on miRNA expressed in the goat mammary gland at peak lactation. The elements generated by this study will now be used as references to decipher the regulation of miRNA expression in the goat mammary gland and to clarify their involvement in the lactation process.
Figures



Similar articles
-
Identification of novel and differentially expressed MicroRNAs of dairy goat mammary gland tissues using solexa sequencing and bioinformatics.PLoS One. 2012;7(11):e49463. doi: 10.1371/journal.pone.0049463. Epub 2012 Nov 14. PLoS One. 2012. PMID: 23166677 Free PMC article.
-
Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology.Mol Biol Rep. 2012 Oct;39(10):9361-71. doi: 10.1007/s11033-012-1779-5. Epub 2012 Jul 5. Mol Biol Rep. 2012. PMID: 22763736
-
Comparative transcriptome profiling of dairy goat microRNAs from dry period and peak lactation mammary gland tissues.PLoS One. 2012;7(12):e52388. doi: 10.1371/journal.pone.0052388. Epub 2012 Dec 26. PLoS One. 2012. PMID: 23300659 Free PMC article.
-
MicroRNA involvement in mammary gland development and breast cancer.Reprod Nutr Dev. 2006 Sep-Oct;46(5):549-56. doi: 10.1051/rnd:2006026. Epub 2006 Sep 23. Reprod Nutr Dev. 2006. PMID: 17107644 Review.
-
Identification and analysis of the expression of microRNA from lactating and nonlactating mammary glands of the Chinese swamp buffalo.J Dairy Sci. 2017 Mar;100(3):1971-1986. doi: 10.3168/jds.2016-11461. Epub 2017 Jan 18. J Dairy Sci. 2017. PMID: 28109598 Review.
Cited by
-
Genome-Wide Association Study towards Genomic Predictive Power for High Production and Quality of Milk in American Alpine Goats.Int J Genomics. 2020 Jul 26;2020:6035694. doi: 10.1155/2020/6035694. eCollection 2020. Int J Genomics. 2020. PMID: 32802828 Free PMC article.
-
The Role of microRNAs in the Mammary Gland Development, Health, and Function of Cattle, Goats, and Sheep.Noncoding RNA. 2021 Dec 13;7(4):78. doi: 10.3390/ncrna7040078. Noncoding RNA. 2021. PMID: 34940759 Free PMC article. Review.
-
Integrative Analysis of Transcriptome and GWAS Data to Identify the Hub Genes Associated With Milk Yield Trait in Buffalo.Front Genet. 2019 Feb 5;10:36. doi: 10.3389/fgene.2019.00036. eCollection 2019. Front Genet. 2019. PMID: 30804981 Free PMC article.
-
Transcriptomic analysis of mammary gland tissues in lactating and non-lactating dairy goats reveals miRNA-mediated regulation of lactation, involution, and remodeling.Front Cell Dev Biol. 2025 May 30;13:1604855. doi: 10.3389/fcell.2025.1604855. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40519264 Free PMC article.
-
High throughput sequencing of small RNAs reveals dynamic microRNAs expression of lipid metabolism during Camellia oleifera and C. meiocarpa seed natural drying.BMC Genomics. 2017 Jul 20;18(1):546. doi: 10.1186/s12864-017-3923-z. BMC Genomics. 2017. PMID: 28728593 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

