Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria
- PMID: 25888176
- PMCID: PMC4475899
- DOI: 10.1128/AEM.00635-15
Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria
Abstract
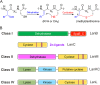
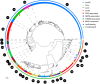
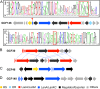
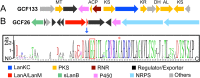
Lanthionine-containing peptides (lanthipeptides) are a rapidly growing family of polycyclic peptide natural products belonging to the large class of ribosomally synthesized and posttranslationally modified peptides (RiPPs). Lanthipeptides are widely distributed in taxonomically distant species, and their currently known biosynthetic systems and biological activities are diverse. Building on the recent natural product gene cluster family (GCF) project, we report here large-scale analysis of lanthipeptide-like biosynthetic gene clusters from Actinobacteria. Our analysis suggests that lanthipeptide biosynthetic pathways, and by extrapolation the natural products themselves, are much more diverse than currently appreciated and contain many different posttranslational modifications. Furthermore, lanthionine synthetases are much more diverse in sequence and domain topology than currently characterized systems, and they are used by the biosynthetic machineries for natural products other than lanthipeptides. The gene cluster families described here significantly expand the chemical diversity and biosynthetic repertoire of lanthionine-related natural products. Biosynthesis of these novel natural products likely involves unusual and unprecedented biochemistries, as illustrated by several examples discussed in this study. In addition, class IV lanthipeptide gene clusters are shown not to be silent, setting the stage to investigate their biological activities.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

