Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP
- PMID: 25888893
- PMCID: PMC4416339
- DOI: 10.1186/s12915-015-0138-0
Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP
Abstract
Background: Recent genome-wide association studies have uncovered genomic loci that underlie an increased risk for atrial fibrillation, the major cardiac arrhythmia in humans. The most significant locus is located in a gene desert at 4q25, approximately 170 kilobases upstream of PITX2, which codes for a transcription factor involved in embryonic left-right asymmetry and cardiac development. However, how this genomic region functionally and structurally relates to PITX2 and atrial fibrillation is unknown.
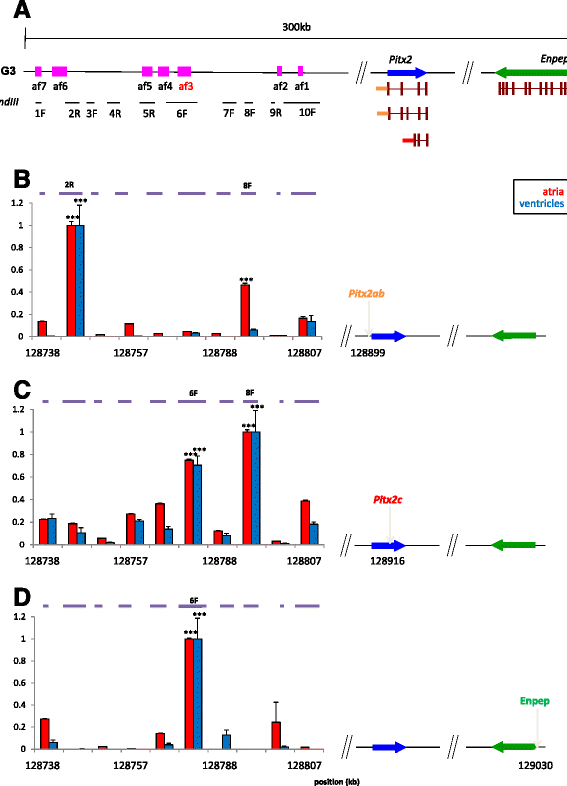
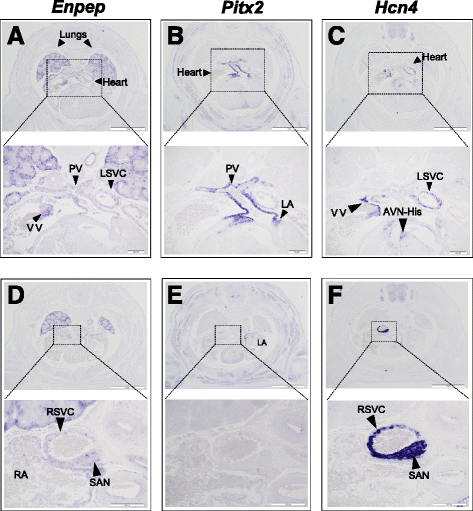
Results: To characterise its function, we tested genomic fragments from 4q25 for transcriptional activity in a mouse atrial cardiomyocyte cell line and in transgenic mouse embryos, identifying a non-tissue-specific potentiator regulatory element. Chromosome conformation capture revealed that this region physically interacts with the promoter of the cardiac specific isoform of Pitx2. Surprisingly, this regulatory region also interacts with the promoter of the next neighbouring gene, Enpep, which we show to be expressed in regions of the developing mouse heart essential for cardiac electrical activity.
Conclusions: Our data suggest that de-regulation of both PITX2 and ENPEP could contribute to an increased risk of atrial fibrillation in carriers of disease-associated variants, and show the challenges that we face in the functional analysis of genome-wide disease associations.
Figures



References
-
- Kirchhof P, Curtis AB, Skanes AC, Gillis AM, Samuel Wann L, John CA. Atrial fibrillation guidelines across the Atlantic: a comparison of the current recommendations of the European Society of Cardiology/European Heart Rhythm Association/European Association of Cardiothoracic Surgeons, the American College of Cardiology Foundation/American Heart Association/Heart Rhythm Society, and the Canadian Cardiovascular Society. Eur Heart J. 2013;34:1471–1474. doi: 10.1093/eurheartj/ehs446. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

