Balanced splicing at the Tat-specific HIV-1 3'ss A3 is critical for HIV-1 replication
- PMID: 25889056
- PMCID: PMC4422144
- DOI: 10.1186/s12977-015-0154-8
Balanced splicing at the Tat-specific HIV-1 3'ss A3 is critical for HIV-1 replication
Abstract
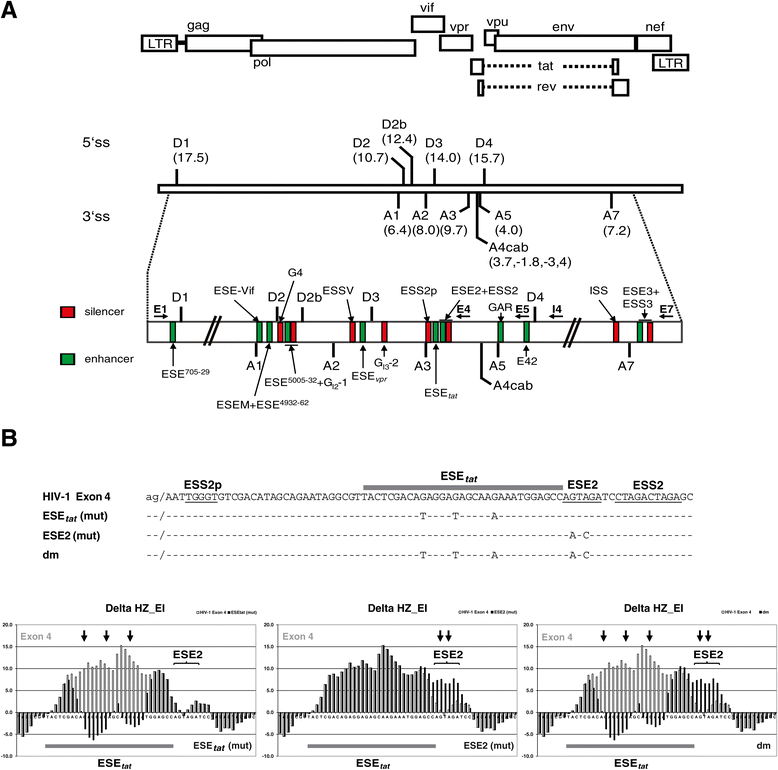
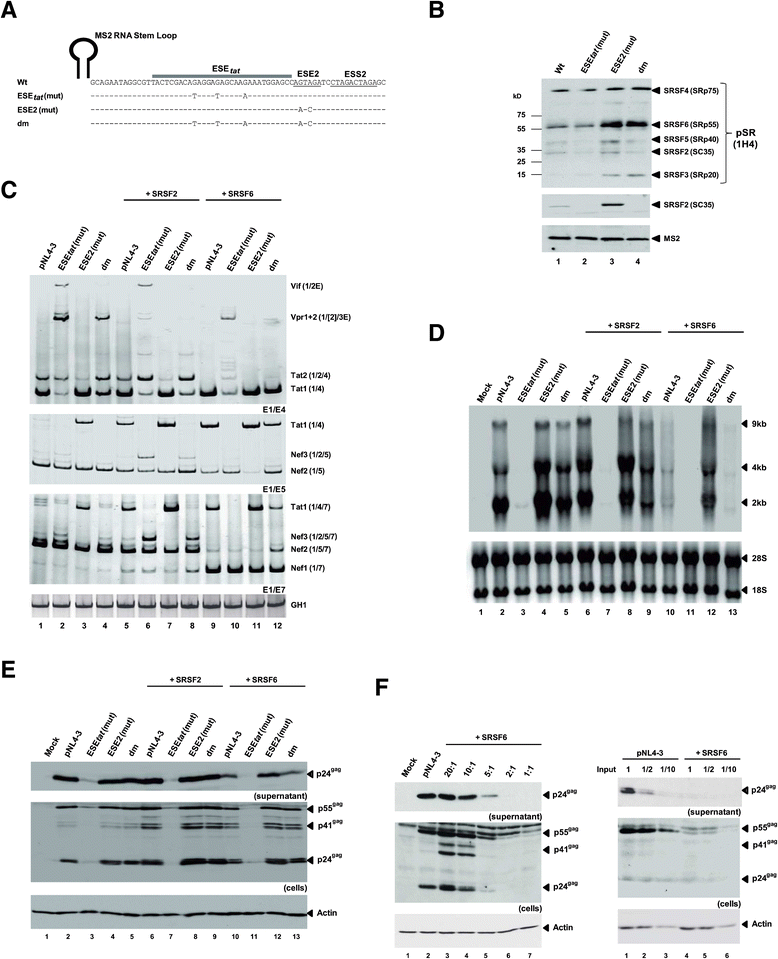
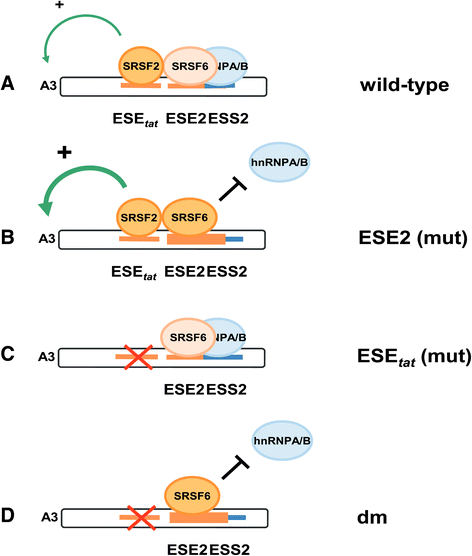
Background: The viral regulatory protein Tat is essential for establishing a productive transcription from the 5'-LTR promoter during the early phase of viral gene expression. Formation of the Tat-encoding mRNAs requires splicing at the viral 3'ss A3, which has previously been shown to be both negatively and positively regulated by the downstream splicing regulatory elements (SREs) ESS2p and ESE2/ESS2. However, using the novel RESCUE-type computational HEXplorer algorithm, we were recently able to identify another splicing enhancer (ESE(5807-5838), henceforth referred to as ESE tat ) located between ESS2p and ESE2/ESS2. Here we show that ESE tat has a great impact on viral tat-mRNA splicing and that it is fundamental for regulated 3'ss A3 usage.
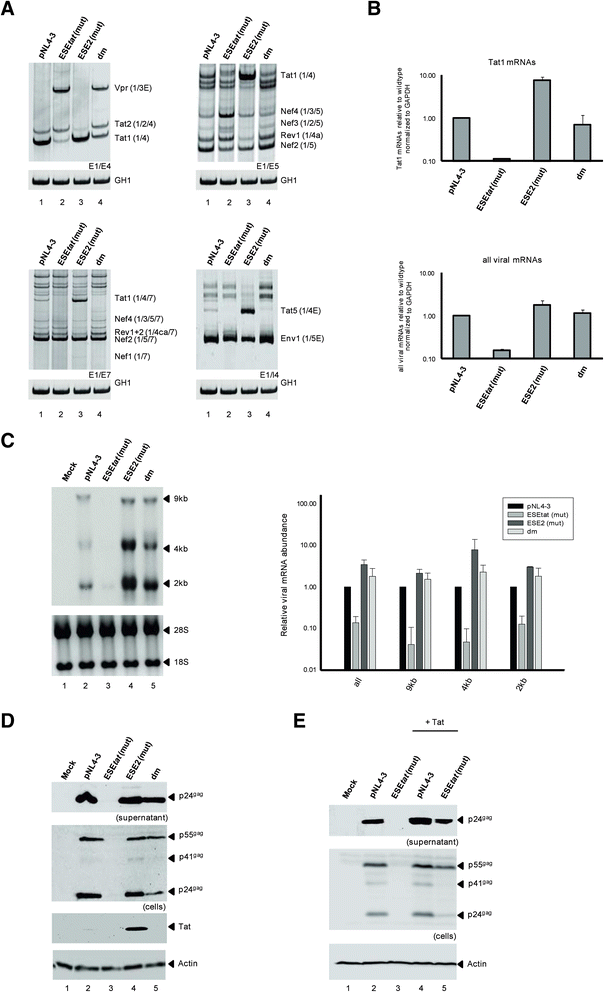
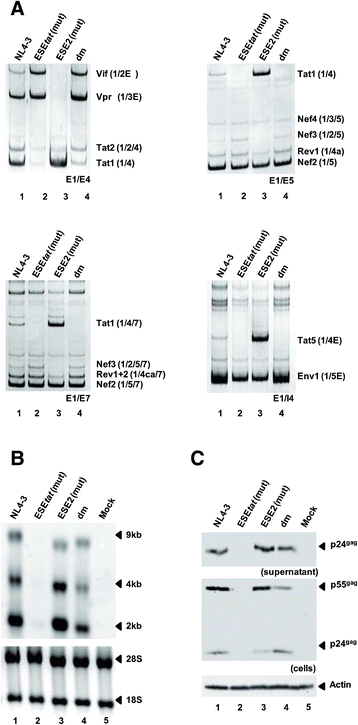
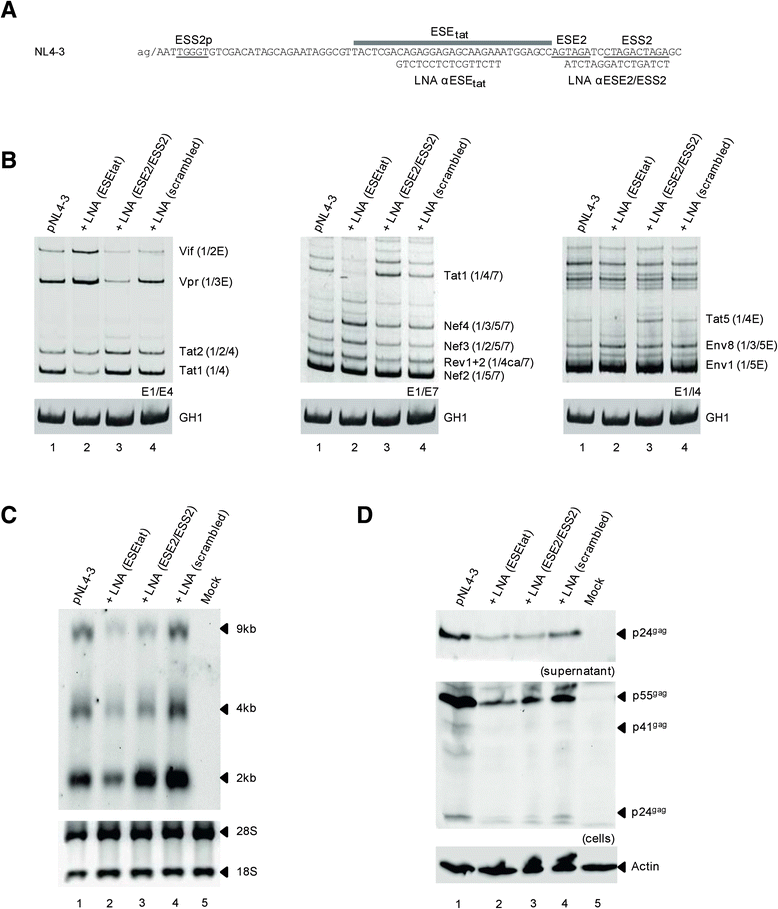
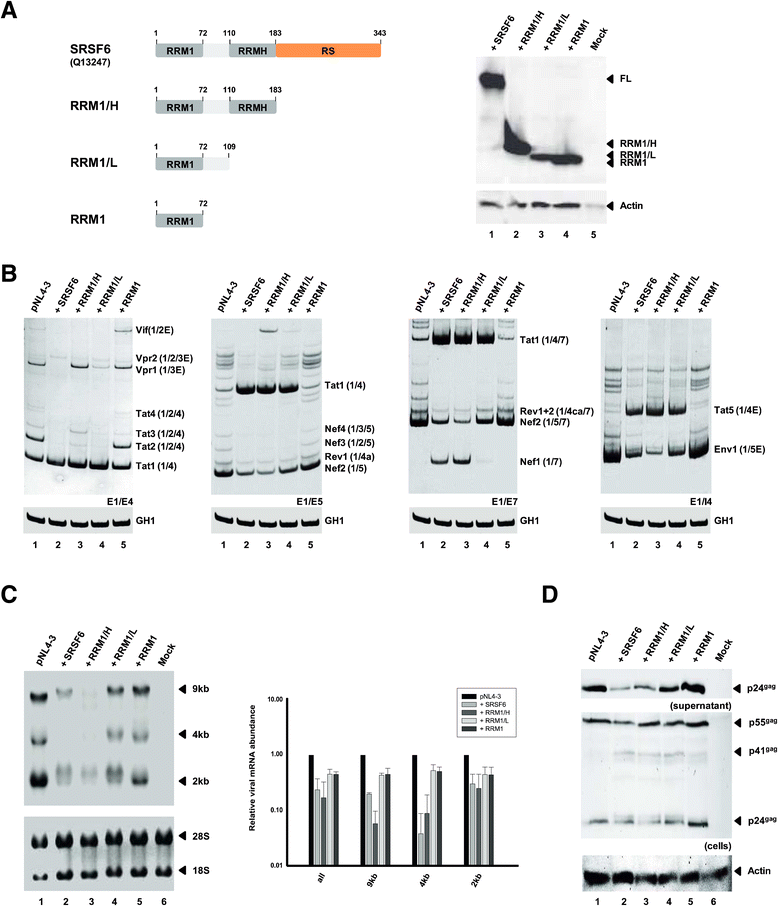
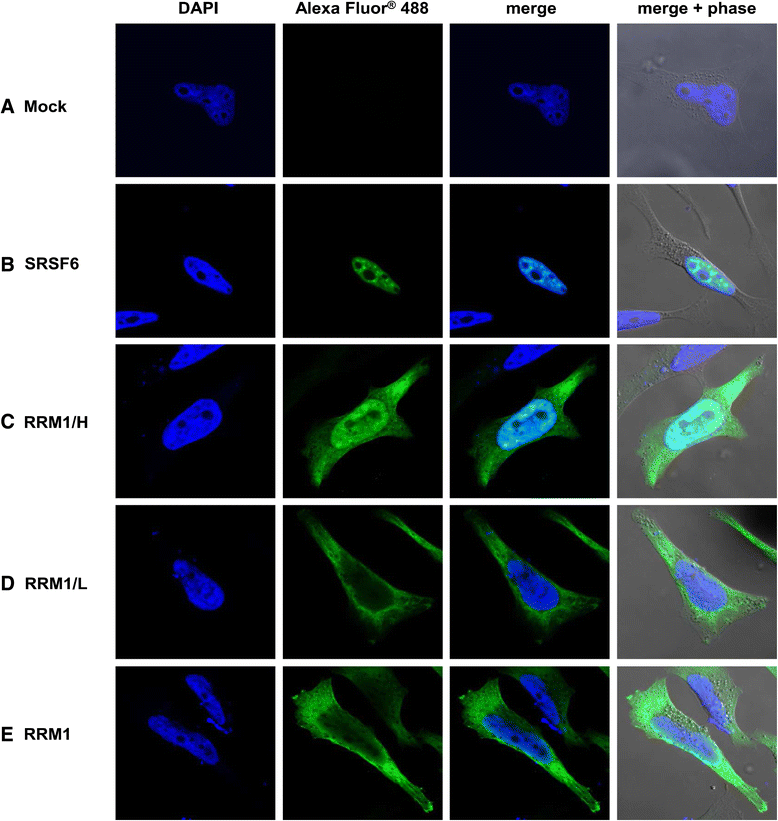
Results: Mutational inactivation or locked nucleic acid (LNA)-directed masking of the ESE tat sequence in the context of a replication-competent virus was associated with a failure (i) to activate viral 3'ss A3 and (ii) to accumulate Tat-encoding mRNA species. Consequently, due to insufficient amounts of Tat protein efficient viral replication was drastically impaired. RNA in vitro binding assays revealed SRSF2 and SRSF6 as candidate splicing factors acting through ESE tat and ESE2 for 3'ss A3 activation. This notion was supported by coexpression experiments, in which wild-type, but not ESE tat -negative provirus responded to higher levels of SRSF2 and SRSF6 proteins with higher levels of tat-mRNA splicing. Remarkably, we could also find that SRSF6 overexpression established an antiviral state within provirus-transfected cells, efficiently blocking virus particle production. For the anti-HIV-1 activity the arginine-serine (RS)-rich domain of the splicing factor was dispensable.
Conclusions: Based on our results, we propose that splicing at 3'ss A3 is dependent on binding of the enhancing SR proteins SRSF2 and SRSF6 to the ESE tat and ESE2 sequence. Mutational inactivation or interference specifically with ESE tat activity by LNA-directed masking seem to account for an early stage defect in viral gene expression, probably by cutting off the supply line of Tat that HIV needs to efficiently transcribe its genome.
Figures
Similar articles
-
A suboptimal 5' splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication.Retrovirology. 2006 Feb 3;3:10. doi: 10.1186/1742-4690-3-10. Retrovirology. 2006. PMID: 16457729 Free PMC article.
-
An exonic splicing silencer downstream of the 3' splice site A2 is required for efficient human immunodeficiency virus type 1 replication.J Virol. 2005 Aug;79(16):10478-86. doi: 10.1128/JVI.79.16.10478-10486.2005. J Virol. 2005. PMID: 16051840 Free PMC article.
-
Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site.J Biol Chem. 2006 Dec 1;281(48):37159-74. doi: 10.1074/jbc.M603864200. Epub 2006 Sep 21. J Biol Chem. 2006. PMID: 16990281
-
Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing.Curr HIV Res. 2006 Jan;4(1):43-55. doi: 10.2174/157016206775197655. Curr HIV Res. 2006. PMID: 16454710 Review.
-
Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication.Adv Virus Res. 2009;74:1-40. doi: 10.1016/S0065-3527(09)74001-1. Adv Virus Res. 2009. PMID: 19698894 Review.
Cited by
-
Transcriptomic Analysis Reveals Key Pathways Influenced by HIV-2 Vpx.Int J Mol Sci. 2025 Apr 8;26(8):3460. doi: 10.3390/ijms26083460. Int J Mol Sci. 2025. PMID: 40331967 Free PMC article.
-
Interferon-Regulated Expression of Cellular Splicing Factors Modulates Multiple Levels of HIV-1 Gene Expression and Replication.Viruses. 2024 Jun 11;16(6):938. doi: 10.3390/v16060938. Viruses. 2024. PMID: 38932230 Free PMC article. Review.
-
Serine/Arginine-rich Splicing Factor 2 Modulates Herpes Simplex Virus Type 1 Replication via Regulating Viral Gene Transcriptional Activity and Pre-mRNA Splicing.J Biol Chem. 2016 Dec 16;291(51):26377-26387. doi: 10.1074/jbc.M116.753046. Epub 2016 Oct 26. J Biol Chem. 2016. PMID: 27784784 Free PMC article.
-
Selective ablation of 3' RNA ends and processive RTs facilitate direct cDNA sequencing of full-length host cell and viral transcripts.Nucleic Acids Res. 2022 Sep 23;50(17):e98. doi: 10.1093/nar/gkac516. Nucleic Acids Res. 2022. PMID: 35736235 Free PMC article.
-
Identification of small molecule modulators of HIV-1 Tat and Rev protein accumulation.Retrovirology. 2017 Jan 26;14(1):7. doi: 10.1186/s12977-017-0330-0. Retrovirology. 2017. PMID: 28122580 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

