GLI2 inhibition abrogates human leukemia stem cell dormancy
- PMID: 25889765
- PMCID: PMC4414375
- DOI: 10.1186/s12967-015-0453-9
GLI2 inhibition abrogates human leukemia stem cell dormancy
Abstract
Background: Dormant leukemia stem cells (LSC) promote therapeutic resistance and leukemic progression as a result of unbridled activation of stem cell gene expression programs. Thus, we hypothesized that 1) deregulation of the hedgehog (Hh) stem cell self-renewal and cell cycle regulatory pathway would promote dormant human LSC generation and 2) that PF-04449913, a clinical antagonist of the GLI2 transcriptional activator, smoothened (SMO), would enhance dormant human LSC eradication.
Methods: To test these postulates, whole transcriptome RNA sequencing (RNA-seq), microarray, qRT-PCR, stromal co-culture, confocal fluorescence microscopic, nanoproteomic, serial transplantation and cell cycle analyses were performed on FACS purified normal, chronic phase (CP) chronic myeloid leukemia (CML), blast crisis (BC) phase CML progenitors with or without PF-04449913 treatment.
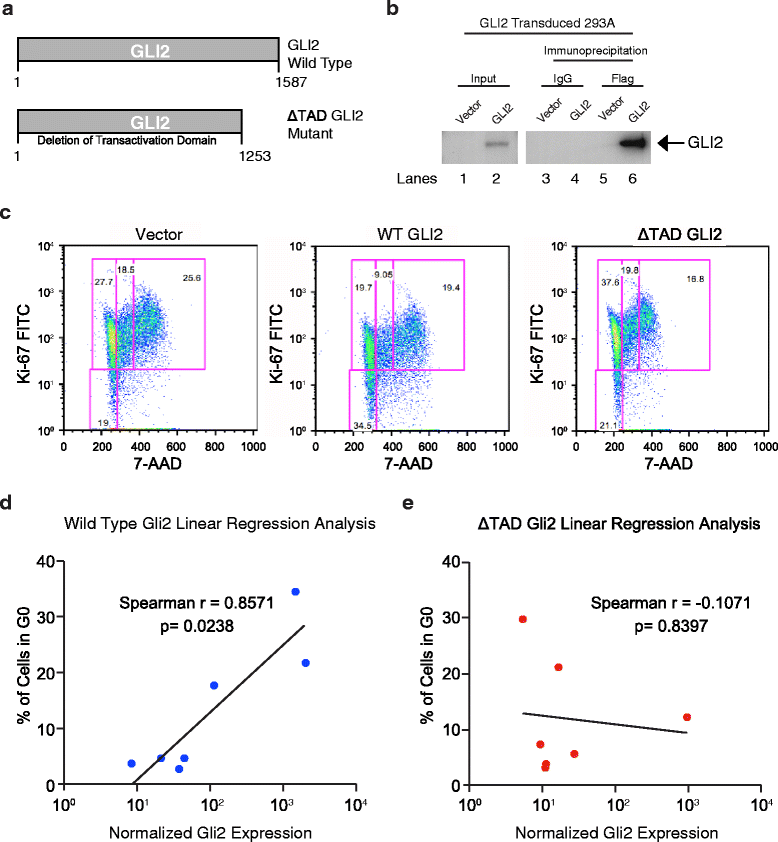
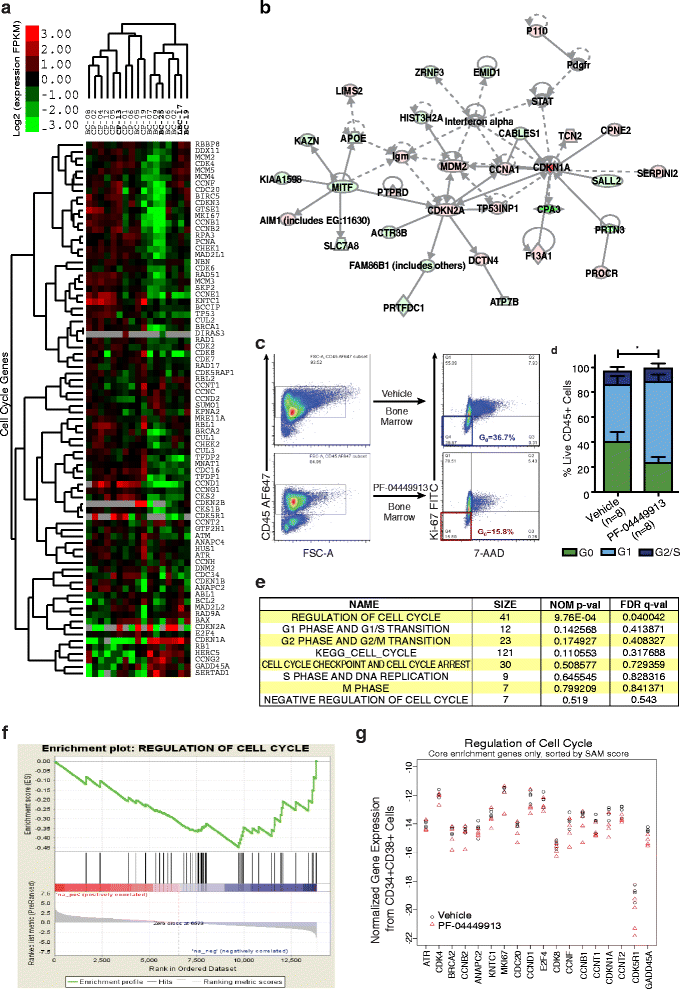
Results: Notably, RNA-seq analyses revealed that Hh pathway and cell cycle regulatory gene overexpression correlated with leukemic progression. While lentivirally enforced GLI2 expression enhanced leukemic progenitor dormancy in stromal co-cultures, this was not observed with a mutant GLI2 lacking a transactivation domain, suggesting that GLI2 expression prevented cell cycle transit. Selective SMO inhibition with PF-04449913 in humanized stromal co-cultures and LSC xenografts reduced downstream GLI2 protein and cell cycle regulatory gene expression. Moreover, SMO inhibition enhanced cell cycle transit and sensitized BC LSC to tyrosine kinase inhibition in vivo at doses that spare normal HSC.
Conclusion: In summary, while GLI2, forms part of a core HH pathway transcriptional regulatory network that promotes human myeloid leukemic progression and dormant LSC generation, selective inhibition with PF-04449913 reduces the dormant LSC burden thereby providing a strong rationale for clinical trials predicated on SMO inhibition in combination with TKIs or chemotherapeutic agents with the ultimate aim of obviating leukemic therapeutic resistance, persistence and progression.
Figures



References
-
- Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94(6):2056–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

