Leveraging Multi-ethnic Evidence for Mapping Complex Traits in Minority Populations: An Empirical Bayes Approach
- PMID: 25892113
- PMCID: PMC4570551
- DOI: 10.1016/j.ajhg.2015.03.008
Leveraging Multi-ethnic Evidence for Mapping Complex Traits in Minority Populations: An Empirical Bayes Approach
Abstract
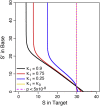
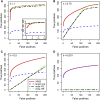
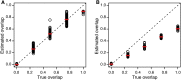
Elucidating the genetic basis of complex traits and diseases in non-European populations is particularly challenging because US minority populations have been under-represented in genetic association studies. We developed an empirical Bayes approach named XPEB (cross-population empirical Bayes), designed to improve the power for mapping complex-trait-associated loci in a minority population by exploiting information from genome-wide association studies (GWASs) from another ethnic population. Taking as input summary statistics from two GWASs-a target GWAS from an ethnic minority population of primary interest and an auxiliary base GWAS (such as a larger GWAS in Europeans)-our XPEB approach reprioritizes SNPs in the target population to compute local false-discovery rates. We demonstrated, through simulations, that whenever the base GWAS harbors relevant information, XPEB gains efficiency. Moreover, XPEB has the ability to discard irrelevant auxiliary information, providing a safeguard against inflated false-discovery rates due to genetic heterogeneity between populations. Applied to a blood-lipids study in African Americans, XPEB more than quadrupled the discoveries from the conventional approach, which used a target GWAS alone, bringing the number of significant loci from 14 to 65. Thus, XPEB offers a flexible framework for mapping complex traits in minority populations.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Coram M.A., Duan Q., Hoffmann T.J., Thornton T., Knowles J.W., Johnson N.A., Ochs-Balcom H.M., Donlon T.A., Martin L.W., Eaton C.B. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 2013;92:904–916. - PMC - PubMed
-
- Chang M.H., Ned R.M., Hong Y., Yesupriya A., Yang Q., Liu T., Janssens A.C., Dowling N.F. Racial/ethnic variation in the association of lipid-related genetic variants with blood lipids in the US adult population. Circ Cardiovasc Genet. 2011;4:523–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268201100001I/HL/NHLBI NIH HHS/United States
- R01 HG006292/HG/NHGRI NIH HHS/United States
- HHSN271201100004C/PHS HHS/United States
- HHSN268201100001C/PHS HHS/United States
- HHSN268201100004I/HL/NHLBI NIH HHS/United States
- HHSN268201100046C/HL/NHLBI NIH HHS/United States
- HHSN268201100003C/WH/WHI NIH HHS/United States
- R01 GM073059/GM/NIGMS NIH HHS/United States
- HHSN268201100003C/PHS HHS/United States
- HHSN268201100004C/PHS HHS/United States
- R01HG006292/HG/NHGRI NIH HHS/United States
- HHSN271201100004C/AG/NIA NIH HHS/United States
- HHSN268201100002C/WH/WHI NIH HHS/United States
- R01HG006124/HG/NHGRI NIH HHS/United States
- R01 HG006124/HG/NHGRI NIH HHS/United States
- HHSN268201100046C/PHS HHS/United States
- HHSN268201100002I/HL/NHLBI NIH HHS/United States
- HHSN268201100002C/PHS HHS/United States
- HHSN268201100001C/WH/WHI NIH HHS/United States
- HHSN268201100004C/WH/WHI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

