Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading
- PMID: 25892223
- PMCID: PMC4445235
- DOI: 10.1016/j.cell.2015.03.012
Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading
Abstract
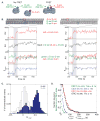
Loading of the ring-shaped Mcm2-7 replicative helicase around DNA licenses eukaryotic origins of replication. During loading, Cdc6, Cdt1, and the origin-recognition complex (ORC) assemble two heterohexameric Mcm2-7 complexes into a head-to-head double hexamer that facilitates bidirectional replication initiation. Using multi-wavelength single-molecule fluorescence to monitor the events of helicase loading, we demonstrate that double-hexamer formation is the result of sequential loading of individual Mcm2-7 complexes. Loading of each Mcm2-7 molecule involves the ordered association and dissociation of distinct Cdc6 and Cdt1 proteins. In contrast, one ORC molecule directs loading of both helicases in each double hexamer. Based on single-molecule FRET, arrival of the second Mcm2-7 results in rapid double-hexamer formation that anticipates Cdc6 and Cdt1 release, suggesting that Mcm-Mcm interactions recruit the second helicase. Our findings reveal the complex protein dynamics that coordinate helicase loading and indicate that distinct mechanisms load the oppositely oriented helicases that are central to bidirectional replication initiation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures






Comment in
-
Single-Molecule Visualization of MCM2-7 DNA Loading: Seeing Is Believing.Cell. 2015 Apr 23;161(3):429-430. doi: 10.1016/j.cell.2015.04.006. Cell. 2015. PMID: 25910200
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

