Global insights into alternative polyadenylation regulation
- PMID: 25892335
- PMCID: PMC4615881
- DOI: 10.1080/15476286.2015.1040974
Global insights into alternative polyadenylation regulation
Abstract
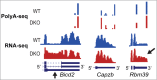
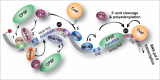
Alternative pre-mRNA processing greatly increases the coding capacity of the human genome and regulatory factors involved in RNA processing play critical roles in tissue development and maintenance. Indeed, abnormal functions of RNA processing factors have been associated with a wide range of human diseases from cancer to neurodegenerative disorders. While many studies have emphasized the importance of alternative splicing (AS), recent high-throughput sequencing efforts have also allowed global surveys of alternative polyadenylation (APA). For the majority of pre-mRNAs, as well as some non-coding transcripts such as lncRNAs, APA selects different 3'-ends and thus modulates the availability of regulatory sites recognized by trans-acting regulatory effectors, including miRs and RNA binding proteins (RBPs). Here, we compare the available technologies for assessing global polyadenylation patterns, summarize the roles of auxiliary factors on APA, and discuss the impact of differential polyA site (pA) selection in the determination of cell fate, transformation and disease.
Keywords: 3′ UTR, 3′-untranslated region; APA, Alternative polyadenylation; AS, Alternative splicing; DM, Myotonic dystrophy; HITS-CLIP, High-throughput sequencing coupled with crosslinking and immunoprecipitation; KD, Knockdown; KO, Knockout; MBNL; PolyA-seq; RBP, RNA binding protein; RNA processing; alternative polyadenylation; microsatellites; myotonic dystrophy; neurological disease; pA, Polyadenylation site.
Figures
References
-
- Shi Y. Alternative polyadenylation: new insights from global analyses. RNA 2012; 18:2105-17; PMID:23097429; http://dx.doi.org/10.1261/rna.035899.112 - DOI - PMC - PubMed
-
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res 2012; 22:1173-83; PMID:22454233; http://dx.doi.org/10.1101/gr.132563.111 - DOI - PMC - PubMed
-
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 2013; 27:2380-96; PMID:24145798; http://dx.doi.org/10.1101/gad.229328.113 - DOI - PMC - PubMed
-
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673-84; PMID:19703394; http://dx.doi.org/10.1016/j.cell.2009.06.016 - DOI - PMC - PubMed
-
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al.. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 2012; 149:538-53; PMID:22502866; http://dx.doi.org/10.1016/j.cell.2012.03.022 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
