XactMice: humanizing mouse bone marrow enables microenvironment reconstitution in a patient-derived xenograft model of head and neck cancer
- PMID: 25893296
- PMCID: PMC4613815
- DOI: 10.1038/onc.2015.94
XactMice: humanizing mouse bone marrow enables microenvironment reconstitution in a patient-derived xenograft model of head and neck cancer
Abstract
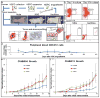
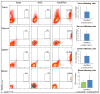
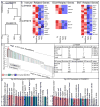
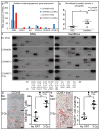
The limitations of cancer cell lines have led to the development of direct patient-derived xenograft models. However, the interplay between the implanted human cancer cells and recruited mouse stromal and immune cells alters the tumor microenvironment and limits the value of these models. To overcome these constraints, we have developed a technique to expand human hematopoietic stem and progenitor cells (HSPCs) and use them to reconstitute the radiation-depleted bone marrow of a NOD/SCID/IL2rg(-/-) (NSG) mouse on which a patient's tumor is then transplanted (XactMice). The human HSPCs produce immune cells that home into the tumor and help replicate its natural microenvironment. Despite previous passage on nude mice, the expression of epithelial, stromal and immune genes in XactMice tumors aligns more closely to that of the patient tumor than to those grown in non-humanized mice-an effect partially facilitated by human cytokines expressed by both the HSPC progeny and the tumor cells. The human immune and stromal cells produced in the XactMice can help recapitulate the microenvironment of an implanted xenograft, reverse the initial genetic drift seen after passage on non-humanized mice and provide a more accurate tumor model to guide patient treatment.
Conflict of interest statement
A.J., Y.R., XJ.W. and D.R.R. are co-inventors of technology presented in this report. The other authors declare no competing financial interests.
Figures


References
-
- Engelholm SA, Vindelov LL, Spang-Thomsen M, Brunner N, Tommerup N, Nielsen MH, et al. Genetic instability of cell lines derived from a single human small cell carcinoma of the lung. European journal of cancer & clinical oncology. 1985;21(7):815–24. Epub 1985/07/01. - PubMed
-
- Hausser HJ, Brenner RE. Phenotypic instability of Saos-2 cells in long-term culture. Biochemical and biophysical research communications. 2005;333(1):216–22. Epub 2005/06/09. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA117802/CA/NCI NIH HHS/United States
- R01 DE015953/DE/NIDCR NIH HHS/United States
- R21 DE019712/DE/NIDCR NIH HHS/United States
- R01 DE024371/DE/NIDCR NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- T32 CA174648/CA/NCI NIH HHS/United States
- R21 CA156114/CA/NCI NIH HHS/United States
- R01 CA117802-06/CA/NCI NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- P30 AR057212/AR/NIAMS NIH HHS/United States
- P30 AR057212-02/AR/NIAMS NIH HHS/United States
- R01 CA149456/CA/NCI NIH HHS/United States
- P01 AI022295/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

