A mass spectrometric-derived cell surface protein atlas
- PMID: 25894527
- PMCID: PMC4404347
- DOI: 10.1371/journal.pone.0121314
A mass spectrometric-derived cell surface protein atlas
Abstract
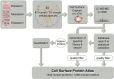
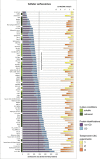
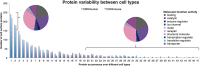
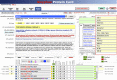
Cell surface proteins are major targets of biomedical research due to their utility as cellular markers and their extracellular accessibility for pharmacological intervention. However, information about the cell surface protein repertoire (the surfaceome) of individual cells is only sparsely available. Here, we applied the Cell Surface Capture (CSC) technology to 41 human and 31 mouse cell types to generate a mass-spectrometry derived Cell Surface Protein Atlas (CSPA) providing cellular surfaceome snapshots at high resolution. The CSPA is presented in form of an easy-to-navigate interactive database, a downloadable data matrix and with tools for targeted surfaceome rediscovery (http://wlab.ethz.ch/cspa). The cellular surfaceome snapshots of different cell types, including cancer cells, resulted in a combined dataset of 1492 human and 1296 mouse cell surface glycoproteins, providing experimental evidence for their cell surface expression on different cell types, including 136 G-protein coupled receptors and 75 membrane receptor tyrosine-protein kinases. Integrated analysis of the CSPA reveals that the concerted biological function of individual cell types is mainly guided by quantitative rather than qualitative surfaceome differences. The CSPA will be useful for the evaluation of drug targets, for the improved classification of cell types and for a better understanding of the surfaceome and its concerted biological functions in complex signaling microenvironments.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

