Reactions of Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase Homologue, with Acetylene and Allene Substrates: Evidence for a Hydration-Dependent Decarboxylation
- PMID: 25894805
- PMCID: PMC4620918
- DOI: 10.1021/acs.biochem.5b00240
Reactions of Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase Homologue, with Acetylene and Allene Substrates: Evidence for a Hydration-Dependent Decarboxylation
Abstract
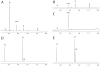
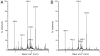
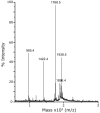
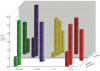
Cg10062 is a cis-3-chloroacrylic acid dehalogenase (cis-CaaD) homologue from Corynebacterium glutamicum with an unknown function and an uninformative genomic context. It shares 53% pairwise sequence similarity with cis-CaaD including the six active site amino acids (Pro-1, His-28, Arg-70, Arg-73, Tyr-103, and Glu-114) that are critical for cis-CaaD activity. However, Cg10062 is a poor cis-CaaD: it lacks catalytic efficiency and isomer specificity. Two acetylene compounds (propiolate and 2-butynoate) and an allene compound, 2,3-butadienoate, were investigated as potential substrates. Cg10062 functions as a hydratase/decarboxylase using propiolate as well as the cis-3-chloro- and 3-bromoacrylates, generating mixtures of malonate semialdehyde and acetaldehyde. The two activities occur sequentially at the active site using the initial substrate. With 2,3-butadienoate and 2-butynoate, Cg10062 functions as a hydratase and converts both to acetoacetate. Mutations of the proposed water-activating residues (E114Q, E114D, and Y103F) have a range of consequences from a reduction in wild type activity to a switch of activities (i.e., hydratase into a hydratase/decarboxylase or vice versa). The intermediates for the hydration and decarboxylation products can be trapped as covalent adducts to Pro-1 when NaCNBH3 is incubated with the E114D mutant and 2,3-butadienoate or 2-butynoate, and the Y103F mutant and 2-butynoate. Three mechanisms are presented to explain these findings. One mechanism involves the direct attack of water on the substrate, whereas the other two mechanisms use covalent catalysis in which a covalent bond forms between Pro-1 and the hydration product or the substrate. The strengths and weaknesses of the mechanisms and the implications for Cg10062 function are discussed.
Figures





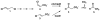
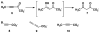

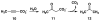





References
-
- Poelarends GJ, Serrano H, Person MD, Johnson WH, Jr., Murzin AG, Whitman CP. Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: Insights into the mechanistic, structural, and evolutionary relationship between isomer-specific 3-chloroacrylic acid dehalogenases. Biochemistry. 2004;43:759–772. - PubMed
-
- Murzin AG. Structural classification of proteins: New superfamilies. Curr. Opin. Struct. Biol. 1996;6:386–394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

